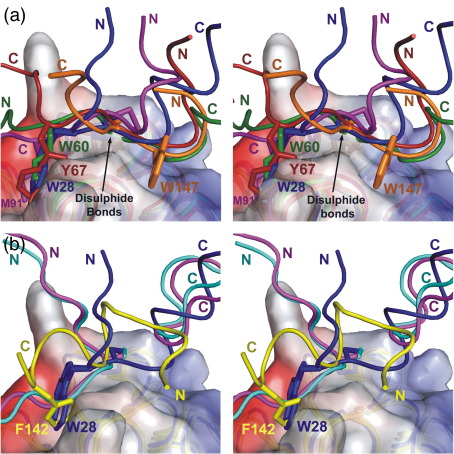
Fig. 5.
Comparison of protein/peptide binding modes in the BsTrxA C32S dimer and other thioredoxin peptide and protein complexes. (a) BsTrxA was superimposed on the thioredoxin from other complexes using PyMOL.35 The superimposed structures, the surface of chain A of BsTrxA, and the peptide or protein chain of the bound molecule are shown. The disulfides and large hydrophobic residues that contribute to binding are also shown. The N- and C-termini of the bound peptides and proteins are indicated with the letters N and C. The proteins shown in (a) are as follows: the BsTrxA dimer with residues 26–38 of chain B shown as cartoons and the side chains of Trp28 and Cys29 shown as sticks (this study; dark blue); the NMR structures of human thioredoxin complexed with substrate peptides from Ref-1 (1CQH; red) or NF-κB (1MDJ; green) with the side chains of residues Cys65 and Tyr67 of the Ref-1 peptide and of residues Trp60 and Cys62 of the NF-κB peptide; the NMR structure of the B. subtilis TrxA and ArsC (2IPT; purple) with residues Cys89 and Met91; and the X-ray structure of the complex between barley thioredoxin and the α-amylase serine proteinase inhibitor (2IWT; orange) with residues Trp147 and Cys148. (b) Comparison of the BsTrxA (dark blue) dimer interface with the binding of thioredoxin reductase by E. coli thioredoxin (1F6M; yellow) and the X-ray structures of the complexes between spinach chloroplast thioredoxins Trx-f and Trx-m and ferrodoxin–thioredoxin reductase (2PU9 and 2PUK; in cyan and magenta, respectively). Trp28 is shown for the TrxA structure (dark blue), and the E. coli thioredoxin reductase structure shows Phe142 (yellow).