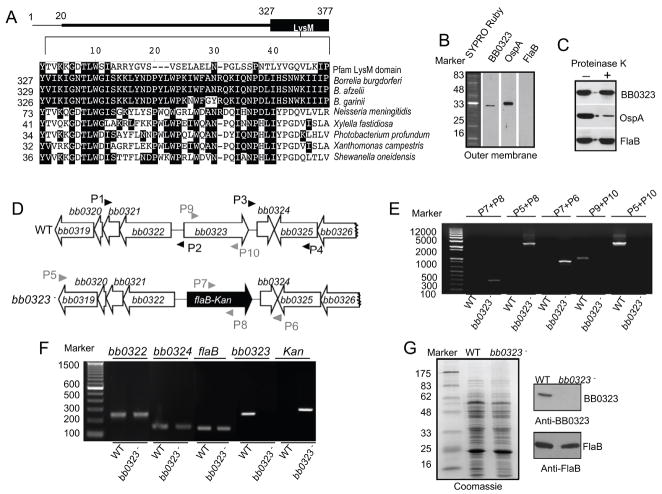
Figure 2. Construction and analysis of the bb0323 mutant B. burgdorferi.
A, Position and amino acid sequence alignment of putative LysM-like domain of B. burgdorferi BB0323. The upper panel represents BB0323 protein indicating an amino terminal signal peptide (encompassing the first 20 amino acids) and a carboxyl terminal LysM domain. Lower panel shows amino acid sequence alignment of LysM-like domain of BB0323 to a designated LysM domain (Pfam accession number PF01476) or to orthologs in major B. burgdorferi sensu lato isolates and representative non-spirochete proteins with closest homologies. Dashes indicate gaps introduced to optimize alignments and identical amino acids are shaded. The numbers on the left are the positions of the amino acid residues in the proteins, for which corresponding species and annotations are indicated in the supplementary text. B, BB0323 is detectable in the outer membrane. Outer membrane vesicles were separated by sucrose density gradient centrifugation, separated by SDS-PAGE, and stained with SYPRO Ruby or immunoblotted with BB0323, OspA and FlaB antibodies. C, BB0323 is partially sensitive to proteinase K-mediated degradation of B. burgdorferi surface proteins. Viable B. burgdorferi were incubated with (+) or without (−) proteinase K for the degradation of protease-sensitive surface proteins and processed for immunoblot analysis using BB0323 antibodies. B. burgdorferi OspA and FlaB antibodies were utilized as controls for surface-exposed and sub-surface proteins, respectively. D, Schematic drawings of the wild type (WT) and the bb0323 mutant (bb0323−) isolates at the bb0323 locus. Genes bb0319 - bb0326 (white box arrows) and the kanamycin-resistance cassette driven by the B. burgdorferi flaB promoter (flaB-Kan, black box arrow) are indicated. Primers P1–P4 (black arrow-heads) were used to amplify 5′ and 3′ arms for homologous recombination, and regions flanking up- and down-stream of the bb0323 locus were ligated on either side of the flaB-Kan cassette as detailed in the text. E, Integration of the mutagenic construct, flab-Kan, in the intended genomic locus. Primers 5–10 (gray arrow-heads, positions indicated in figure 2D) were used for PCR analysis using isolated DNA from wild type (WT) or mutant B. burgdorferi (bb0323−) and subjected to gel electrophoresis. The combination of primers used for PCR is indicated at the top, and migration of the DNA ladder is shown on the left. F, RT-PCR assessment of bb0323 ablation and the polar effects of mutagenesis. Total RNA was isolated from wild type B. burgdorferi (WT) or bb0323 mutant (bb0323−) converted to cDNA and used to amplify regions within bb0323, flaB, kanamycin and genes surrounding bb0323 locus (bb0322 and bb0324) and visualized on a gel. G, Protein analysis of wild type and mutant isolates. Equal amounts of protein were by SDS-PAGE gel, and either stained with Coomassie blue (left panel) or immunoblotted with BB0323 and FlaB antibodies (right panel). Migration of protein standards is shown to the left in kDa.