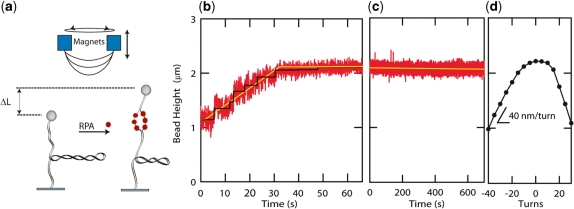
Figure 2.
hRPA dsDNA binding and helix unwinding. (a) Schematic of the experimental scheme. hRPA binds to ssDNA that is transiently exposed upon helix breathing. Further protein binding unwinds the helix. Unwinding leads to removal of negative supercoils that were previously applied to the molecule (n=−40). (b) Height of the magnetic bead versus time after introduction of hRPA. The increasing height signals hRPA binding. The binding curve reveals a reaction that progresses linearly in time (yellow line, guide to the eye). Step-like behavior in the binding traces indicates single enzyme binding events (black line, fit based on step finding algorithm). The reaction saturates when the length of the molecule reaches the length of the non-supercoiled molecule. (c) After removal of the unbound hRPA, hRPA remains stably bound to the ssDNA-bubble ([NaCl]=30 mM). (d) Rotation curve of molecule taken prior to the reaction. The slope of the rotation curve at experimental reaction conditions [NaCl]=30 mM; F=0.55 pN equals 40 nm/turn. A singly bound RPA molecule is expected to unwind the helix by ∼3 turns, thus leading to a length increase of 120 nm.