Figure 4.
Evaluation of the reweighting scheme. (a and b) Unadjusted and re-weighted base-level counts for reads from the WT experiment mapped to the sense strand of a 1-kb coding region in S. cerevisiae (YOL086C). The graey bars near the x-axis indicate unmappable genomic locations. (c) The 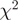 goodness-of-fit statistics based on unadjusted and reweighted counts for 552 highly expressed regions of constant expression. (d) Smoothed histograms of the reduction in
goodness-of-fit statistics based on unadjusted and reweighted counts for 552 highly expressed regions of constant expression. (d) Smoothed histograms of the reduction in 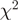 goodness-of-fit statistics when using the re-weighting scheme, evaluated in five different experiments. Values greater than zero indicate that the re-weighting scheme improves the uniformity of the read distribution.
goodness-of-fit statistics when using the re-weighting scheme, evaluated in five different experiments. Values greater than zero indicate that the re-weighting scheme improves the uniformity of the read distribution.

