Abstract
Efavirenz (EFV) is a nonnucleoside reverse transcriptase inhibitor (NNRTI) of HIV-1 reverse transcriptase (RT) used for the treatment of AIDS. RT is a heterodimer composed of p66 and p51 subunits; p51 is produced from p66 by C-terminal truncation by HIV protease. The monomers can form p66/p66 and p51/p51 homodimers as well as p66/p51 heterodimer. Dimerization and efavirenz binding are coupled processes. In the crystal structure of the p66/p51—EFV complex, the drug is bound to the p66 subunit. The binding of efavirenz to wild-type and dimerization-defective RT proteins was studied by equilibrium dialysis, tryptophan fluorescence and native gel electrophoresis. A 1:1 binding stoichiometry was determined for both monomers and homodimers. Equilibrium dissociation constants are ~2.5 μM for both p66— and p51—EFV complexes, 250 nM for p66/p66—EFV complex, and 7 nM for p51/p51—EFV complex. An equilibrium dissociation constant of 92 nM for p66/p51—EFV complex was calculated from the thermodynamic linkage between dimerization and inhibitor binding. Binding and unbinding kinetics monitored by fluorescence were slow. Progress curve analyses revealed a one-step, direct binding mechanism with association rate constants k1 ~13.5 M–1 s–1 for monomers and heterodimer and dissociation rate constants k–1 ~1 × 10–4 s–1 for monomers. A conformational selection mechanism is proposed to account for the slow association rate. These results show that efavirenz is a slow, tight-binding inhibitor capable of binding all forms of RT and suggest that the NNRTI binding site in monomers and dimers is similar.
HIV-11 RT converts single-stranded viral RNA into double-stranded proviral DNA. The enzyme has two activities, DNA polymerase and RNase H. The biologically relevant form is a heterodimer composed of two subunits, p66 and p51 (1). The subunits are products of the same gene and have identical N-terminal amino acid sequences; p51 lacks the C-terminal RNase H domain (2-4). The individual subunits can also form homodimers. The p66 subunit in the heterodimer has both polymerase and RNase H active sites (5). The monomeric species are devoid of enzymatic activity (3, 4). Due to its essential role in the HIV lifecycle, RT is a major target of antiretroviral drugs (6). Two classes of inhibitors have been developed and approved for clinical use, NRTIs and NNRTIs. The NNRTIs are highly effective and relatively noncytotoxic (7). These small, amphiphilic, noncompetitive inhibitors nestle into a hydrophobic pocket ~10 Å away from the polymerase active site in the p66 subunit of RT (8, 9). NNRTIs primarily interfere with reverse transcription, but they also affect late stages of HIV replication in Gag-Pol polyprotein processing (10-12).
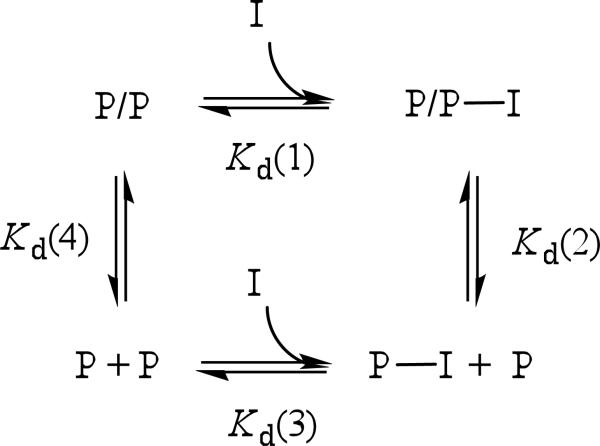
NNRTIs have diverse effects on RT subunit dimerization. Efavirenz (EFV) and nevirapine (NVP) enhance subunit interactions (13, 14), delavirdine has little or no effect (13), and TSAOe3T, BBNH and BBSH weaken subunit interactions (15, 16). The evidence for these results derives from multiple techniques including yeast two-hybrid, pull-down assays, urea induced dissociation, size exclusion chromatography, and sedimentation equilibrium studies. To explain the contrasting effects of NNRTI binding on RT, we previously proposed a thermodynamic cycle (14). In Scheme 1, P denotes p66 or p51 monomer, P/P is p66/p51 heterodimer, p66/p66 homodimer, or p51/p51 homodimer, and I is NNRTI. The thermodynamic linkage between NNRTI binding and RT subunit dimerization makes the following predictions (1) NNRTIs bind to both monomeric and dimeric forms of RT proteins. The crystal structures of RT—NNRTI complexes show one drug bound per heterodimer (8, 9). In solution, the stoichiometry of drug binding to dimer is not known. (2) NNRTIs that enhance dimerization bind more tightly to dimers. Conversely, NNRTIs that weaken dimerization bind more tightly to monomers. Identifying and quantifying the various protein-ligand interactions is essential for thorough understanding of the inhibition mechanism of NNRTIs. The previous thermodynamic cycle (Scheme 1 in ref 14) makes the additional prediction that low concentrations of inhibitor will promote dimerization if Kd(1) < Kd(3).2 However, eventually Le Châtelier's principle will shift the equilibrium towards the formation of P—I at high concentrations of inhibitor.
Scheme 1.
Thermodynamic Linkage of NNRTI Binding and Subunit Dimerization
Previous sedimentation equilibrium studies showed that efavirenz enhances the formation of p66/p51, p66/p66, and p51/p51 by 25-, 50-, and 600-fold (14). Here we measure the binding of efavirenz to p66 and p51 monomers in wild-type and dimerization-defective mutant RTs and determine the binding stoichiometry of monomers and homodimers. Binding stoichiometry and equilibrium dissociation constants for drug binding to dimer and monomer, Kd(1) and Kd(3), were determined by equilibrium dialysis. The kinetics of drug binding to monomers and heterodimer were monitored by intrinsic protein fluorescence. Finally, the binding of [14C] efavirenz to p66 monomer and p66/p66 homodimer was visualized by Blue Native gel electrophoresis.
EXPERIMENTAL PROCEDURES
Materials
Efavirenz was obtained from the NIH AIDS Research and Reference Reagent Program (Germantown, MD). [14C] efavirenz (Specific Activity: 52 mCi/mmol) was purchased from Vitrax (Placentia, CA). Dialysis tubing was purchased from Spectrum Labs (Rancho Dominguez, CA). Rapid Equilibrium Dialysis (RED) Device and TCEP were purchased from Pierce (Rockford, IL). Econo-Safe scintillation fluid was purchased from Atlantic Nuclear Corporation (Canton, MA). Oligodeoxynucleotide primers, 5% Coomassie blue G-250 sample additive, and NativePAGE Novex Bis-Tris gel system were purchased from Invitrogen Corp. (Carlsbad, CA). EZ-Run Protein Gel Staining solution was purchased from Fisher Scientific (Fair Lawn, NJ). Biochemical reagents were purchased from Roche Applied Science (Indianapolis, IN). Other chemicals were from Sigma Chemicals (St. Louis, MO). RT buffer D is 0.05 M Tris (pH 7.0), 25 mM NaCl, 1 mM EDTA, and 10% (v/v) glycerol.
Protein Preparation
HIV-1 RT proteins with N-terminal hexahistadine extensions were expressed in Escherichia coli M15 strains containing plasmid p6H RT for p66, p6H RT51 for p51, or p6H RT-PR for p66/p51 heterodimer and purified by Ni–NTA, S-Sepharose, and DEAE chromatography as previously described (14, 17). Protein concentration is determined from absorbance at 280 nm and is expressed in monomer units (14, 18). Protein stock solutions were dialyzed overnight into RT buffer D containing 1 mM TCEP3 prior to use.
Dimerization-defective RT proteins were prepared from plasmids p6H RT and p6H RT51 containing the W401A mutation (19). The W401A mutation was introduced by one round of mutagenesis using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotide primer sequences were: forward, 5’-GGGAAACAGCGTGGCCAGAGTATTGGCAAGCCACCTG-3’; reverse, 5’-CAGGTGGCTTGCCAATACTCTGTCCACGCTGTTTCCC-3’. All mutations were confirmed by DNA sequencing at Agencourt Bioscience (Beverly, MA).
Equilibrium Dialysis
Equilibrium dialysis experiments were conducted using 1.5 mL RNase/DNase free amber microcentrifuge tubes and 4-mm dialysis tubing with 3,500 molecular weight cut off or RED devices. A 1 mM stock solution of [14C] efavirenz in DMF was prepared. A 250 μL aliquot of RT solution was loaded into dialysis tubing or one chamber of the RED device. RT concentrations were 0.1–10 μM p51, 1–10 μM p51W401A, 2–4 μM p51L234A, 0.4–5 μM p66, and 0.8–7.5 μM p66W401A. RT buffer D containing 1 mM TCEP and 0.2–20 μM [14C] efavirenz was used as dialysate buffer. For microcentrifuge tubes, the dialysis bag and 1 mL of dialysate buffer were placed in the tube and the tube was capped. For RED devices, 0.4 mL of dialysate buffer was placed in the other chamber. The samples were set up in triplicate, secured to a benchtop rotator, and dialyzed at 4 °C. Wild-type RT proteins were dialyzed for up to 5 days; W401A mutant proteins were dialyzed for 30 h. Equilibration of efavirenz across the membrane occured by 20 h.
Efavirenz binding was quantified by counting three 50 μL aliquots of the inside protein solution and outside dialysate solution in 5 mL of scintillation fluid using a Beckman Coulter LS6500 Multi purpose scintillation counter. A buffer blank and 50 μL aliquots of the initial dialysate solution were also counted. Bound ligand concentration was calculated from:
| (1) |
where [Iin] is the total concentration of free and bound efavirenz inside the dialysis tubing or RED chamber, and [Iout] = [I] is the concentration of free efavirenz in the outside dialysate. Scintillation counting data were converted to molarity and fit to mathematical models in the Dialfit program as described in the Appendix. The value of Ka(4) was fixed in the data analysis using ln Ka = 8.3 for p51/p51 homodimer and ln Ka = 12.4 for p66/p66 homodimer (14).
Isothermal Titration Calorimetry
ITC experiments were performed on a Microcal VP-ITC microcalorimeter. Wild-type p51 solutions (1.5 and 3.0 μM) were titrated with efavirenz (200 μM) in RT buffer D containing 3% DMF at 5 °C. Prior to the reaction p51 was dialyzed into RT buffer D containing 3% DMF to eliminate any solvent effects. Aliquots of 5, 10, and 15 μL of the efavirenz solution were added over 60 min to a final concentration of 40 μM. The amounts of heat released after each addition of efavirenz into the p51 solution and the buffer blank were identical, indicating that (1) the binding event is too slow to measure by this technique or (2) ΔΗ = 0.
Fluorescence
Absorbance was measured on a Cary 3E UV–vis spectrophotometer. Fluorescence was measured on a PC1 photon counting spectrofluorometer (ISS, Champaign, IL) in ratio mode under magic angle conditions using 4 nm excitation and 16 nm emission bandwidths at 5 °C. The sample compartment was flushed with nitrogen to prevent condensation. Samples were placed in 45 μL quartz cells with 3-mm path length (Starna Cells, Inc., Atascadero, CA). Absorbance at 280 nm was < 0.3 to avoid inner filter effects. Fluorescence quantum yields Φ were measured at 295 nm excitation wavelength relative to NATA in water with Φ = 0.23 at 5 °C. The quantum yield of NATA at 5 °C was determined relative to tryptophan in water at 295 nm excitation wavelength, 25 °C, with Φ = 0.14 (20).
Association and dissociation kinetics of RT proteins and efavirenz were monitored by fluorescence using Vinci 1.6.SP7 software (ISS, Champaign, IL). Intrinsic tryptophan fluorescence was measured at 295 nm excitation wavelength, 340 nm emission wavelength using NATA in water as reference. Slow kinetic intensity data were collected from samples and NATA every 30 s (signal averaged over 5 s) for 4–5 h, then every 5 min (signal averaged over 10 s) for 27 h. Fluorescence intensity F = Is / Ir was calculated from the ratio of sample intensity Is to reference intensity Ir to correct for instrumental drift.
Association reactions were started by adding 2 μL of a diluted efavirenz stock solution (250 mM in DMF) to 80 μL of 2.5 μM p66W401A or p51W401A, 4.5 μM wild-type p51, or 10-fold dilution of 20 μM p66/p51 (85% dimer). The solution was mixed in the cell for 5 s and immediately placed in the fluorometer. Final efavirenz concentrations were 5–40 μM. Dissociation reactions were started by 100-fold dilution of 20 μM p66W401A or p51W401A equilibrated with 35 μM efavirenz. The change in intrinsic tryptophan fluorescence due to binding or unbinding of efavirenz was fit to a single exponential function.
| (2a) |
| (2b) |
where F(t) is intensity at time t, F0 is intensity at t = 0, F∞ is intensity of the last time point, and the Cs are constants.
Native Gel Electrophoresis
BN-PAGE was carried out using the Novex Bis-Tris gel system as described previously (21). A 5–10 μL aliquot of 2 μM p66W401A and 0.8–5 μM p66 in the absence or presence of NNRTI was mixed with 0.3 μL Coomassie G-250 sample additive, 2.5 μL NativePAGE Sample Buffer, and water to a final volume of 15 μL. Gels were stained with EZ-Run Protein Gel Staining solution and destained in water. For gels containing [14C] efavirenz, p66 was incubated for 2 h or 1 week and subjected to BN-PAGE. Gels were imaged by a PhosphorImager (Amersham Biosciences, Piscataway, NJ), viewed with ImageQuant software, and then stained in EZ-Run Protein Gel Staining solution and destained in water.
RESULTS
Equilibrium Dialysis
Binding of efavirenz to p66 and p51 is coupled to formation of homo- and heterodimers (Scheme 1). Dimerization constants in the absence and presence of NNRTI are characterized by Kd(4) and Kd(2), while inhibitor dissociation constants of dimer and monomer complexes are Kd(1) and Kd(3). Dimerization constants for p66/p66 and p51/p51 homodimers in the absence and presence of efavirenz were previously determined by sedimentation equilibrium (14). Equilibrium dialysis was used to determine inhibitor dissociation constants Kd(1) and Kd(3).
Equilibrium binding experiments were initially set up with p51, because the dimerization constants in the absence and presence of efavirenz, Kd(4) = 230 μM for p51/p51 and Kd(2) = 0.37 μM for p51/p51—I, provide access to both monomer and homodimer. The first binding experiments used 10 μM p51 (7.5% homodimer) and 20 μM [14C] efavirenz. Dialysis was terminated and samples were analyzed at 30 h and at 3, 5 and 7 days. After 30 h the ratio of efavirenz to p51 was ~0.84:1, indicating a binding stoichiometry of either one inhibitor per p51 monomer or two inhibitors per p51/p51 homodimer. The ratio of efavirenz to p51 decreased to 0.68:1 after 3 days, 0.52:1 after 5 days, and 0.49:1 after 7 days. A ratio of one efavirenz per p51/p51 homodimer is consistent with the stoichiometry in the crystal structure of p66/p51—EFV complex (9). Due to the slow dimerization all experiments examining efavirenz binding to dimeric species were allowed to bind for 5 days. To confirm that the 30 h dialysis with wild-type p51 represents efavirenz binding to monomer, equilibrium dialysis experiments were also performed using dimerization-defective RT proteins. Two dimerization-defective mutations reported in the literature are L234A (22, 23) and W401A (19). L234A is a primer grip mutation; W401A is a mutation in the tryptophan repeat motif of the connection subdomain. The presence of either of these mutations in the p66 or p51 subunit of the heterodimer result in dimerization deficiency, the mutation in p66 having the most detrimental effect. Equilibrium dialysis experiments set up with 3–6 μM p51L234A and 5–12 μM [14C] efavirenz failed to detect any bound efavirenz. Thus the L234A mutation prevents not only dimerization but also efavirenz binding. This is not surprising given that L234 is a contact residue in the NNRTI binding pocket.
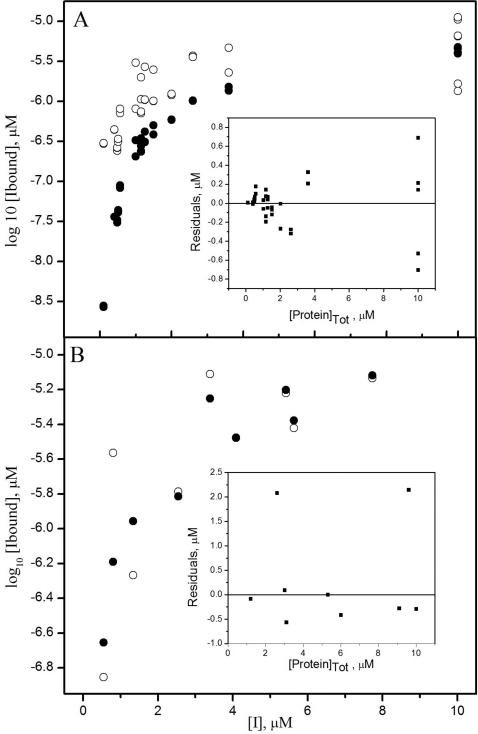
Inhibitor dissociation constants Kd(1) and Kd(3) were determined by simultaneously varying protein and efavirenz concentrations in equilibrium dialysis experiments. The data sets for multiple concentrations of protein and efavirenz were analyzed with the Dialfit program (Appendix). The ln Ka(4) value, where Ka(4) is the equilibrium association constant of p51/p51 or p66/p66 homodimers (14), is set as a constant and ln Ka values for inhibitor binding to monomers and dimers are allowed to float. Data sets for wild-type RT proteins equilibrated with efavirenz were fit to the coupled equilibria in Scheme 1; data sets for the dimerization-defective mutants were fit neglecting the dimerization reaction. Weighted least square fits were performed until the fits converged. Figure 1A shows efavirenz binding data for wild-type p51 together with the fit to eqs A1a – A1c for the coupled equilibria. The log of [Ibound] is plotted for clarity; [Ibound] in μM was used in the data analysis. To illustrate the range of efavirenz and protein concentrations used in the experiments, the residuals [Ibound]exp – [Ibound]calc are plotted versus total protein concentration (inset). Figure 1B shows efavirenz binding data for p51W401A and the fit to eq A1a for a simple binding equilibrium.
Figure 1.
Equilibrium dialysis data for p51. (A) wild-type p51 equilibrated with efavirenz for 5 days: (○) experimental values [Ibound]exp, and (•) calculated values [Ibound]calc from Dialfit using eqs A1a–Alc. (B) p51W401A equilibrated with efavirenz for 30 h: (○) [Ibound]exp, and (•) [Ibound]calc from Dialfit using eq A1a. Insets show residuals ([Ibound]exp – [Ibound]calc) versus total protein concentration [Protein]Tot in each measurement.
Table 1 gives the results of the global analyses for wild-type and dimerization-defective RT proteins. The dissociation constants Kd(1) and Kd(3) for efavirenz binding to dimers and monomers were calculated from Ka values with Kd = 1/Ka. The inhibitor dissociation constants Kd(1) of wild-type homodimer—EFV complexes is about 36-fold tighter for p51/p51—EFV than p66/p66—EFV: Kd(p51/p51—I) = 7 nM compared to Kd(p66/p66—I) = 250 nM. The inhibitor dissociation constants Kd(3) of wild-type and dimerization-defective monomer—EFV complexes are much weaker, in the μM range. The Kd(3) values for wild-type p66 and p51 measured after equilibration with efavirenz for 5 days are inaccurate. At high protein concentrations the free monomer concentration is too low to detect, and at low protein concentrations binding of efavirenz to monomer is too weak to detect. The data set for wild-type p51 equilibrated for 30 h with efavirenz provides a more reliable value for Kd(p51—I) = 1.7 μM, because at 30 h wild-type p51 is ~70% monomers. The inhibitor dissociation constants Kd(3) for p51W401A—EFV and p66W401A—EFV of 2.4–2.7 μM are within error of the value for wild-type p51, suggesting that the efavirenz binding site is the same in all the monomers.
Table 1.
Dissociation Constantsa
| protein | dialysis time | ln Ka(1) | Kd (1), μM | ln Ka(3) | Kd (3), μM |
|---|---|---|---|---|---|
| p51 | 30 h | 17.3 ± 0.6 | 0.030 (0.017 – 0.056) |
13.3 ± 0.6 | 1.7 (0.95 – 3.2) |
| p51 | 5 days | 18.9 ± 0.8 | 0.0068 (0.0028 – 0.014) |
10 ± 1 | 28 (10.1 – 75.0) |
| p51W401A | 30 h | 12.9 ± 0.6 | 2.5 (1.3 – 4.4) |
||
| p66 | 5 days | 15.2 ± 0.5 | 0.25 (0.15 – 0.41) |
11 ± 1 | 19 (5.6 – 61) |
| p66W401A | 30 h | 12.8 ± 0.3 | 2.7 (2.0 – 3.6) |
RT buffer D, 1 mM TCEP, at 5 °C. Global analysis of data sets for each protein and dialysis time from Dialfit. Errors are 95% confidence intervals.
Binding Kinetics
RT contains multiple tryptophan residues, 19 in p66 and 18 in p51 (Figure 2). Tryptophan fluorescence is exquisitely sensitive to the local electrostatic environment of the indole chromophore (24, 25). Changes in RT fluorescence associated with dimerization and NNRTI binding have been reported (26, 27). The fluorescence changes due to dimerization were attributed to the tryptophan repeat motif in the connection subdomain spanning residues 398–414. The NNRTI binding pocket contains β-sheet β12–β13–β14, which has two tryptophans W229 and W239; W229 is in the loop between β-strands 12 and 13 and W239 is in β-strand 14. These two tryptophans may report conformational changes upon inhibitor binding.
Figure 2.
Structure of HIV-1 RT complexed with efavirenz (1FK9): p66 (purple), p51 (green), efavirenz (cyan), and tryptophans (red).
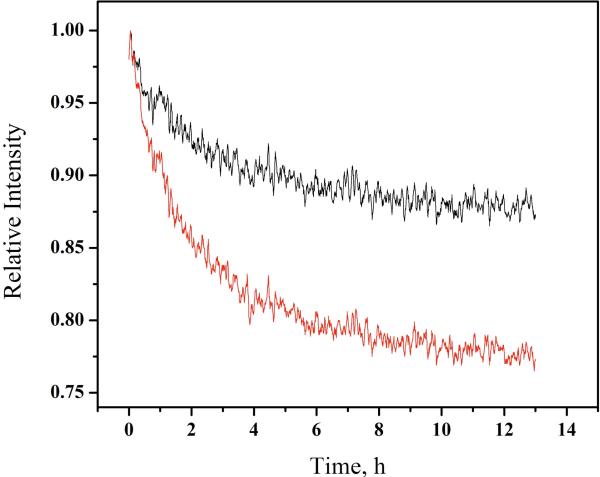
The kinetics of inhibitor binding to RT proteins were monitored by tryptophan fluorescence. Figure 3 shows that efavirenz binds slowly to p51 monomer and p66/p51 heterodimer. About 50% of the overall fluorescence change occurs in ~2 h. In order to measure heterodimer fluorescence, a 20 μM solution containing 83% dimer was diluted 10-fold and efavirenz was added immediately to start the kinetics experiment before dissociation of the dimer occurs (t1/2 = 2 days; 28). The intensity change for the heterodimer is about half that of the monomer, consistent with an effect on tryptophan residues in only one subunit.
Figure 3.
Association of efavirenz to (—) p66/p51 and ( ) p51 monitored by tryptophan fluorescence at 5 °C. λex = 295 nm, λem = 340 nm. 20 μM p66/p51 (83% heterodimer) diluted to 2 μM prior to adding efavirenz; 4.5 μM p51 (97% monomer). [EFV]:[protein] 2:1.
) p51 monitored by tryptophan fluorescence at 5 °C. λex = 295 nm, λem = 340 nm. 20 μM p66/p51 (83% heterodimer) diluted to 2 μM prior to adding efavirenz; 4.5 μM p51 (97% monomer). [EFV]:[protein] 2:1.
Two kinetic mechanisms have been used to account for the slow binding of inhibitors to enzymes (Scheme 2; 29, 30). Mechanism A depicts direct binding of inhibitor I to enzyme E, where the association and dissociation rate constants k1 and k–1 are inherently slow. Mechanism B depicts an induced-fit model with fast equilibration of inhibitor and enzyme to form an intermediate complex EI, followed by slow isomerization of EI complex to form the final complex EI*. To discriminate between the two mechanisms, the observed rate constant kobs from the progress curve of the enzyme reaction, is determined as a function of inhibitor concentration. A plot of kobs versus inhibitor concentration is linear for Mechanism A and hyperbolic for Mechanism B.
Scheme 2.
Mechanisms of Slow Binding Inhibition
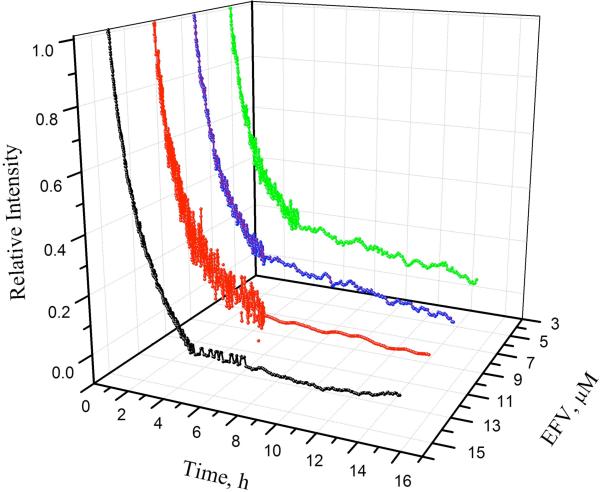
Progress curves for efavirenz binding to p66W401A, p51W401A, and p66/p51 were measured at multiple inhibitor concentrations. Figure 4 shows the set of curves for p51W401A. The solid lines are the fits to eq 2a to obtain values of kobs. Similar curves were obtained for p66W401A and p66/p51. Figure 5 shows the plots of kobs versus inhibitor concentration. The linear fits are consistent with Mechanism A, where
| (3) |
The values of the rate constants k1 and k–1 calculated from the slopes and intercepts of Figure 5 are given in Table 2. All three proteins have similar association rate constants k1 ≈ 13.5 M–1s–1. Additionally, the dissociation rate constants k–1 were about 5.9–8.1 × 10–5 s–1, corresponding to t1/2 ≈ 2.7 h. Having defined the binding modality of efavirenz, values of Kd(3)app were calculated from the ratio of k–1/k1.
FIGURE 4.
Progress curves for p51W401A binding to efavirenz monitored by tryptophan fluorescence at 5 °C. λex = 295 nm, λem = 340 nm. 2.5 μM p51W401A; ( ) 5 μM, (
) 5 μM, ( ) 8 μM, (
) 8 μM, ( ) 11 μM, and (—) 14 μM efavirenz. Data acquired at 30 s intervals for first 4–5 h, then at 5 min intervals. Data were fit to eq 2a to obtain kobs.
) 11 μM, and (—) 14 μM efavirenz. Data acquired at 30 s intervals for first 4–5 h, then at 5 min intervals. Data were fit to eq 2a to obtain kobs.
FIGURE 5.
Progress curve analysis of efavirenz binding to (▲) p66/p51, ( ) p51W401A, (
) p51W401A, ( ) p66W401A. Lines are fits to eq 3. Error bars give the range of average values from 2 experiments.
) p66W401A. Lines are fits to eq 3. Error bars give the range of average values from 2 experiments.
Table 2.
Kinetic Parametersa
The kinetics of dissociation of p66W401A— and p51W401A—EFV complexes were measured under essentially irreversible conditions, so that at equilibrium < 3% of monomer—EFV complex is present (Figure 6). Dissociation rate constants k–1(diss) ≈ 9.0 × 10–5 s–1, or t1/2 ≈ 2.1 h, were obtained for both monomers from fitting the data to eq 2b. The k–1(diss) values from kinetics measurements are close to the k–1 values determined from the plots of kobs versus [I] (Table 2). The equilibrium dissociation constants Kd(3) calculated from the ratio of the rate constants k–1(diss)/k1 for monomer binding are 2.5-fold higher than the value determined by equilibrium dialysis.
FIGURE 6.
Dissociation of ( ) p51W401A—EFV and (
) p51W401A—EFV and ( ) p66W401A—EFV complexes monitored by tryptophan fluorescence at 5 °C. λex = 295 nm, λem = 340 nm. Black curves are fits to eq 2b. Data acquired at 30 s intervals for first 4–5 h, then at 5 min intervals.
) p66W401A—EFV complexes monitored by tryptophan fluorescence at 5 °C. λex = 295 nm, λem = 340 nm. Black curves are fits to eq 2b. Data acquired at 30 s intervals for first 4–5 h, then at 5 min intervals.
Fluorescence Quantum Yields
Fluorescence quantum yields of dimerization-defective monomers and wild-type heterodimer were measured in the absence and presence of efavirenz. To measure the extent of quenching, most of the protein must be bound to efavirenz. The quantum yields of monomer—EFV complexes were measured on solutions containing 1 μM monomer and 30 μM efavirenz equilibrated for 30 h at 5 °C, giving 94% monomer—EFV complex. The quantum yields of p66W401A and p51 W401A monomers are the same within error (Table 3). Efavirenz binding decreases the quantum yield of both monomers by a factor of 3. In order to measure the quantum yield of the heterodimer, a 20 μM solution containing 83% dimer was diluted 50-fold and scanned immediately as above. The quantum yield of the heterodimer is approximately 20% lower than that of the monomers. The quantum yield of p66/p51—EFV complex was measured on solutions containing 20 μM p66/p51 and 40 μM efavirenz equilibrated for 1 wk at 5 °C prior to dilution. Because efavirenz enhances dimerization 25-fold and binds more tightly to dimer than monomer, this solution contains 98% p66/p51—EFV complex. Efavirenz binding to heterodimer only quenches the fluorescence by a factor of 1.6.
Table 3.
Quantum Yieldsa
| Protein | Φ |
|---|---|
| p51W401A | 0.14 ± 0.01 |
| + efavirenz | 0.05 ± 0.02 |
| p66W401A | 0.15 ± 0.01 |
| + efavirenz | 0.05 ± 0.02 |
| p66/p51 | 0.11 ± 0.01 |
| + efavirenz | 0.07 ± 0.01 |
| NATA | 0.23 |
RT buffer D, 1 mM TCEP, at 5 °C, λex = 295 nm. Errors are standard deviations of 4 experiments.
Native Gel Electrophoresis
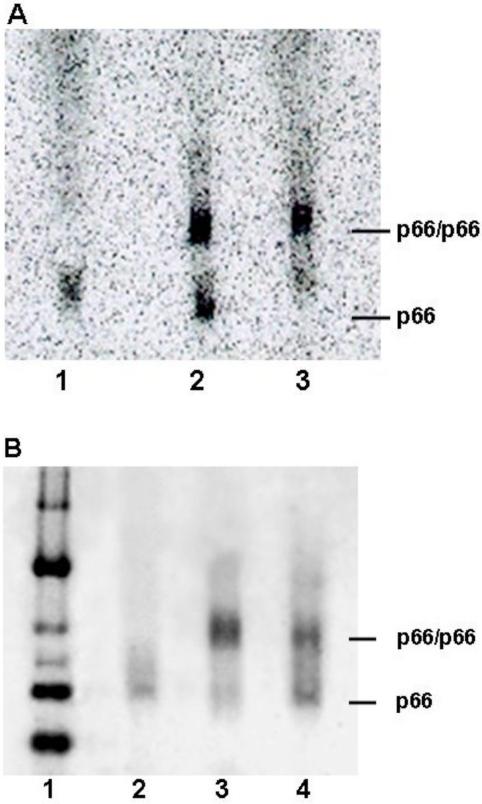
BN-PAGE has been used to monitor dimerization of RT proteins in the absence and presence of efavirenz (21). The slow dissociation rate of efavirenz (t1/2 ~2 h) makes it possible to visualize binding of [14C] efavirenz to monomer on gels. The p66W401A and wild-type p66 were incubated with a 0.7:1.0 ratio of [14C] efavirenz to protein. The wild-type p66 concentration was 5 μM or approximately 53% homodimer. Lane 1 shows [14C] efavirenz binding to p66W401A monomer. Lane 2 shows [14C] efavirenz binding to the mixture of wild-type p66 monomer and p66/p66 homodimer. Lastly, Lane 3 shows enhancement of dimerization by efavirenz after equilibration of wild-type p66 with [14C] and excess cold efavirenz for 1 wk, giving 91% p66/p66—EFV complex. Thus BN-PAGE supports the conclusions from equilibrium dialysis that efavirenz binds RT monomers as well as homodimers.
Nevirapine has been reported to have disparate effects on RT dimerization. Yeast-two hybrid experiments indicate small enhancement of dimerization, whereas urea denaturation studies find no effect (16, 23). BN-PAGE was performed using p66 incubated with excess efavirenz or nevirapine for 1 wk. Figure 7B shows that both NNRTIs enhance dimerization with efavirenz having the greater effect. These results are consistent with the findings by yeast-two hybrid.
FIGURE 7.
Blue Native polyacrylamide gel electrophoresis of p66 in the absence and presence of NNRTIs. (A) Monomer and homodimer binding to [14C] EFV: (lane 1) 2 μM p66W401A incubated 2 h with [14C] EFV, (lane 2) 5 μM p66 incubated 2 h with [14C] EFV, and (lane 3) 5 μM p66 incubated 1 wk with [14C] EFV. (B) wild-type p66 incubated 1 wk in the absence and presence of NNRTI 1 wk: (lane 1) Native Markers; (lane 2) 0.8 μM p66; (lane 3) 3 μM p66, 25 μM EFV; and (lane 4) 3 μM p66, 25 μM NVP.
DISCUSSION
Although RT has been extensively studied for almost two decades, new functions continue to be discovered for this enigmatic enzyme. This paper reports two novel functions: (1) efavirenz, and presumably also other NNRTIs, binds to monomeric forms of RT and (2) efavirenz is a slow binding inhibitor of monomers and heterodimer. The biological significance of monomer binding is presently unknown. NNRTIs have been found to affect both early and late stages of the HIV-1 replication cycle by multiple mechanisms (31, 32). Efavirenz interacts at the level of reverse transcription by inhibiting DNA polymerase activity, enhancing polymerase-dependent RNase H activity (3'-DNA directed) and partially inhibiting polymerase-independent RNase H activity (5'-RNA directed). It also inhibits plus-strand initiation by affecting the ability of RT to bind the RNA polypurine tract. During late stages of HIV-1 replication, efavirenz enhances processing and homodimerization of a 90 kDa Pol polyprotein in a yeast two-hybrid system and increases intracellular processing of Gag and Gag-Pol precursor polyproteins in HIV-1 transfected cells (11). By increasing the processing of these polyproteins, efavirenz lowers viral production due to decreased levels of full constructs for incorporation into a budding particle. Essential to the above processes is defining the binding properties of the species, whether monomer or dimer, to which efavirenz binds. Drug design requires an immense understanding of the target. This study suggests that monomeric forms of RT may be potential targets for HIV-1 therapeutics. It also sparks development of high throughput screening assays based on p66 and p51 monomers to evaluate binding of new drugs to wild-type and drug resistance mutant RTs.
The two crystal structures of RT—EFV complexes show 1:1 binding stoichiometry (9, 33). Currently no crystal structures are available for homodimers or monomers of RT. Equilibrium dialysis indicated a 1:1 stoichiometry for p66/p66— and p51/p51—EFV complexes. A 1:1 binding stoichiometry for monomer—EFV complexes was also obtained by equilibrium dialysis for wild-type p51 and dimerization-deficient p66W401A and p51W401A, albeit with a lower affinity than the homodimers (Table 1). The apparent free energies of efavirenz binding to homodimers at 5 °C, ΔG278 = –RTlnKa, are –35.1 kJ/mol for p66/p66—I and –43.6 kJ/mol for p51/p51—I. The more favorable binding energy of the p51/p51 homodimer may be attributed to better contacts between efavirenz and the protein or more facile formation of the binding pocket in a dimer lacking 2 RNase H domains. All monomers have similar efavirenz binding energies, ΔG278 ~ –30 kJ/mol, which is 5–12 kJ/mol less favorable than binding to homodimers.
The dissociation constants Kd(1) and Kd(3) from equilibrium dialysis (Table 1) and Kd(2) and Kd(4) from previous sedimentation equilibrium experiments, allow us to complete the thermodynamic linkage of NNRTI binding and subunit dimerization proposed for RT (14). In the closed cycle of Scheme 1, ΔG = 0. Substituting ΔG278 gives
| (4) |
In the case of p66, the left side of eq 4 sums to –68 ± 6 kJ/mol and the right side to –64 ± 4 kJ/mol. In the case of p51, the left side of eq 4 totals –64 ± 4 kJ/mol and the right side –63 ± 6 kJ/mol. These results for homodimers confirm the hypothesis that NNRTI binding is coupled to subunit dimerization. Thus we can then calculate the dissociation constant of the p66/p51—EFV complex Kd(1) from the cycle in Scheme 1, where Kd(2) and Kd(4) are the dissociation constants of the heterodimer in the presence and absence of efavirenz (14), and Kd(3) is the dissociation constant of p66— or p51—EFV complexes. Rearranging eq 4 gives RT lnKa(1) = 38 ± 6 kJ/mol or Kd(1) = 92 ± 5 nM. Most studies of efavirenz binding to RT have employed polymerase activity assays, carried out in the presence of template/primer and dNTP (12, 34, 35). Maga et al. (35) reported a dissociation constant of 150 nM for free RT—EFV complex extracted from enzymatic data. Geitmann et al. (36) measured binding of several NNRTIs to immobilized wild-type and drug resistance mutant RTs by SPR at 25 °C in buffer containing 0.005% surfactant and 3% (v/v) DMSO. An overall dissociation constant of 45 nM was obtained for efavirenz binding to wild-type RT.
The binding kinetics monitored by tryptophan fluorescence establish that efavirenz is a slow, tight binding inhibitor of RT. Dissociation rate constants k—1(diss) of 9.0 × 10–5 s–1, or t1/2 = 2.2 h, were obtained for monomer—EFV complexes. Association rate constants k1 of 13.5 M–1s–1 were obtained for a reversible, direct binding reaction of efavirenz to both monomers and heterodimer (Scheme 2, Mechanism A). This suggests that NNRTI binding occurs by an analogous process for monomeric and dimeric species of RT, despite the difference in efavirenz binding affinity. Slow binding inhibitors described in the literature follow either direct binding or conformational change inhibition models (Scheme 2). For example, small azasugar inhibitors of β-glucosidase and yeast isomaltase bind by the direct Mechanism A with association rate constants ranging from 23 M–1 s–1 to 7.3 × 104 M–1 s–1 (37). These inhibitors also have slow dissociation rate constants of 0.16–6.7 × 10–2 s–1. By contrast, peptide α-ketoacid analogues are slow binding inhibitors of hepatitis C virus NS3 protease that bind by the induced-fit Mechanism B (39). These inhibitors undergo rapid equilibration with the enzyme in the first step of EI complex formation with k1 = 6.5 × 107 M–1 s–1 and k–1 = 0.2 s–1. The subsequent step is a slow isomerization to EI* with k2 = 1.7–7.5 × 10–3 s–1 and k–2 = 0.57–1.8 × 10–5 s–1 or t1/2 = 11–48 h.
A few previous reports noted slow onset of inhibition by NNRTIs. For example, 5–20 min pre-incubation periods of RT with NNRTIs were required to witness inhibition of polymerase and RNase H activity (31, 35, 39, 40). The slow association rate constants reported here (Table 2) could be caused by a conformational selection step involving exclusive binding of a lowly populated conformer of either protein or inhibitor. The chemical structure of efavirenz is provided in Figure 2. The benzoxazinone ring system is rigid with free rotation of the cyclopropyl ethynyl group (41). Efavirenz is in the same position in the binding pocket in both crystal structures of wild-type RT—EFV complex. However, the cyclopropyl ethynyl group is rotated ~100° in the drug resistance mutant K103N RT—EFV complex relative to the position in the wild-type structures (9). The rigidity of the efavirenz core together with the ability of the binding pocket to accommodate different orientations of the cyclopropyl ethynyl group excludes conformational selection of the inhibitor as the culprit.
A selected-fit model has been proposed in which a conformational pre-equilibration of the protein precedes inhibitor binding (36, 42). In this model for the slow binding, efavirenz would bind preferentially to a less populated conformer of RT proteins. Productive collisions occurring between inhibitor and this conformer would induce a slow shift in the conformational equilibrium favoring formation of the binding pocket and subsequently the EI complex. A less likely alternative would be severe orientation effects resulting in unproductive collisions of E and I that slow formation of EI complex in the direct binding Mechanism A. A selected-fit model gave the best fit to the SPR data for wild-type RT and efavirenz with an overall association rate constant kon = 5.5 × 104 M–1 s–1. This kon value is 3 orders of magnitude faster then our association rate constant k1 for efavirenz binding to p66/p51. The dissociation rate constant koff ≈ 2.3 × 10–3 s–1 is about 20-fold faster then the k–1 value obtained from progress curve analysis. A probable explanation for the faster binding kinetics is the different solution conditions used in the SPR assay. Osmolytes such as detergents, organic solvents, and salts affect protein solution structure and interactions (43, 44).
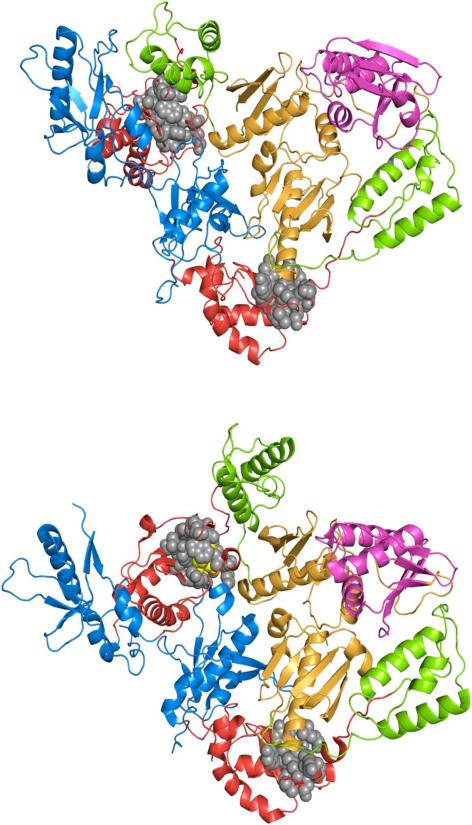
The NNRTI binding pocket is absent from structures of RT and RT—substrate complexes (45, 46). The polymerase domain is composed of four subdomains: fingers, palm, thumb, and connection. The structure of RT is asymmetric and the subdomains are in different orientations in the p66 and p51 subunits (Figure 8, upper; 45). In RT–NNRTI complexes, the binding pocket is in the palm of the p66 subunit of the heterodimer (Figure 8, lower; 9). LPC software identifies 14 residues that contact efavirenz in the NNRTI binding pocket of the p66 subunit: L100, K101, K103, V106, V179, Y181, Y188, G190, F227, W229, L234, H235, P236 and Y318 (47). The efavirenz contact residues are highlighted in Figure 8 to illustrate the location of these residues in the two subunits. Although the contact residues are clustered in both subunits, these residues do not form a binding pocket in the p51 subunit. The p66/p66 and p51/p51 homodimers may also have asymmetric structures, because they both have polymerase activity (3). Additionally the homodimers probably have similar NNRTI binding pockets in the subunit that binds the inhibitor, because p66 and p51 have identical amino acid sequences and similar folding patterns of the polymerase subdomains. Given that p66 and p51 monomers are capable of forming a competent NNRTI binding pocket, presumably the polymerase domain of both monomers must adopt a conformation analogous to that of the p66 subunit in the heterodimer.
FIGURE 8.
Structures of HIV-1 (upper) RT (1DLO) and (lower) RT—EFV complex (1FK9). Polymerase domains: fingers (blue), palm (red), thumb (green), and connection (orange) subdomains; RNase H domain (magenta). Efavirenz (yellow) and contact residues (grey) are shown using the van der Waals radii.
ACKNOWLEDGEMENT
The authors are grateful to Ms. Kathryn J. Howard for suggesting the gel experiment with [14C] efavirenz and for advice about site-directed mutagenesis and protein purification. We also thank Dr. Clare Woodward for suggesting that NNRTIs bind to monomers and Drs. Vernon Anderson and Jonathan Karn for helpful discussions.
APPENDIX
Dialfit: Mathematical Models and Data Analysis Leslie A Holladay §
There are two different ways in which the model equations may be written: Case A and Case B.
Case A
For Case A the following equilibrium constants are defined as:
| (A1a) |
| (A1b) |
| (A1c) |
The equilibrium dialysis experiment involves computing the total molar concentration of inhibitor bound to both monomer and dimer [Ibound], by subtracting the concentration of free inhibitor outside the dialysis bag [Iout] = [I] from the total concentration of inhibitor inside the bag [Iin] as in eq 1. For the data analysis, the observable variable we wish to model is [Ibound]. The total concentration of protein [P]tot inside the bag is presumed to be known to higher precision than that of the free and bound inhibitor concentrations. [Ibound] is the observable to be predicted knowing [I] and [P]tot along with the current estimates for Ka(1), Ka(3), and Ka(4). Dialfit is applicable to any experiment that provides data for [Ibound] and [I], knowing [P]tot.
To compute [Ibound], the concentration of free protein monomer [P] must first be computed. The conservation of mass equation is
| (A2) |
Rearranging and substituting terms with the equilibrium constants from eqs A1 results in a quadratic in [P],
| (A3a) |
where
| (A3b) |
| (A3c) |
| (A3d) |
The only physical meaningful root of eq (A3a) is the positive one. The fitting function may be written as
| (A4) |
The values of [Ibound] determined over a wide range of total inhibitor and total protein concentrations are globally fitted to eq A4. Note that Ka(1) and Ka(4) cannot be separated and thus one value must be a fixed parameter.
Case B
For Case B two equilibrium constants are defined in eqs (A1a) and (A1b); the third equilibrium constant is defined as
| (A5) |
Here too [Ibound] is the observable to be predicted knowing [I] and [P]tot, but with the current estimates for Ka(2), Ka(3), and Ka(4). The conservation of mass eq (A2) and quadratic eqs (A3a), (A3c), and (A3d) are the same as in Case A; eq (A3b) becomes
| (A6) |
Again, the physically meaningful root of eq (A3a) is the positive one. The fitting function is
| (A7) |
Note that Cases A and B are mathematically equivalent from the relationship Ka(1) Ka(4) = Ka(2) Ka(3). The two parameters Ka(2) and Ka(3) are not separable, and thus the value of Ka(2) must be fixed.
Weighted Least Squares and Parameter Standard Errors
The global data sets have a very wide range of values for [Ibound] and [I]. For radioactive counts, the relative standard deviation is equal to , where N is the number of counts (49). The relative variance is equal to 1/N. [Ibound] is computed from the difference in counts inside and outside the bag. Define the number of counts inside the bag as Ni and the number of counts outside the bag as No. Then the relative variance of the difference (Ni – No) = (Ni + No) / (NiNo). In the situation in which the values for [Ibound] and [I] vary over several orders of magnitude, it is essential to use weighted least squares because the variance will also vary over a wide range (50). The weight Wj of any (Ni – No)j value is the reciprocal of the variance, Wj = (NiNo) / (Ni +No). The actual weights used are normalized so that the Σj Wj = 1 to cause the returned residual error in the fit to be correct. It is clear from in Figure 1 (insets) that the errors in [Ibound] are very heteroscedastic, The errors in the fitted variable vary a lot with respect to the independent variable. Only if the errors are homoscedastic is unweighted least squares appropriate.
Standard errors for the parameter values were computed using the “balanced bootstrap” with 100 trials (51, 52). If any of the individual bootstrap trials was more than three standard deviations away from the parameter estimate that trial was deleted as an outlier and the standard deviation recomputed. The 95% confidence intervals are computed using the standard deviation from the estimated parameter value.
Footnotes
This work was supported by NIH grant GM071267.
Abbreviations: BBNH, N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone; BBSH, (4-tert-butylbenzoyl)-2-hydroxy-1-salicylyl hydrazone; BN-PAGE, Blue Native polyacrylamide gel electrophoresis; DMF, dimethyl formamide; DMSO, dimethylsulfoxide; EDTA, ethylenediaminetetraacetic acid; EFV, efavirenz; HIV-1, human immunodeficiency virus type 1; ITC, isothermal titration calorimetry; NATA, N-acetyltryptophanamide; Ni–NTA, nitriloacetic acid; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NVP, nevirapine; PR, HIV-1 protease; RT, reverse transcriptase; SPR, surface plasmon resonance; Tris, tris(hydroxymethyl)aminomethane; TCEP, tris(2-carboxyethyl)phosphine; TSAOe3T, 1-(spiro{4”-amino-2”,2”-dioxo-1”,2”-oxathiole-5”,3′-[2′,5′-bis-O-(tert-butyldimethylsilyl)-β-d-ribofuranosyl]})-3-ethylthymine.
In the previous thermodynamic cycle, reaction 2 for dimerization in the presence of NNRTI is written as P/P—I + I = 2 P—I. The dissociation constant for this reaction is a composite equilibrium constant equal to Kd(2) / Kd(3).
Addition of 1 mM TCEP to RT buffer D lowered the pH from 7.0 to 6.5.
REFERENCES
- 1.di Marzo Veronese F, Copeland TD, DeVico AL, Rahman R, Oroszlan S, Gallo RC, Sarngadharan MG. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 2.Hizi A, McGill C, Hughes SH. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Restle T, Muller B, Goody RS. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 4.Restle T, Muller B, Goody RS. RNase H activity of HIV reverse transcriptase is confined exclusively to the dimeric forms. FEBS Lett. 1992;300:97–100. doi: 10.1016/0014-5793(92)80172-d. [DOI] [PubMed] [Google Scholar]
- 5.Le Grice SFJ, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai CH, Lee PY, Stollar V, Li ML. Antiviral therapy targeting viral polymerase. Curr. Pharm. Des. 2006;12:1339–1355. doi: 10.2174/138161206776361156. [DOI] [PubMed] [Google Scholar]
- 7.De Clercq E. The role of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Antiviral Res. 1998;38:153–179. doi: 10.1016/s0166-3542(98)00025-4. [DOI] [PubMed] [Google Scholar]
- 8.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 9.Ren J, Milton J, Weaver KL, Short SA, Stuart DI, Stammers DK. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure. 2000;8:1089–1094. doi: 10.1016/s0969-2126(00)00513-x. [DOI] [PubMed] [Google Scholar]
- 10.Merluzzi VJ, Hargrave KD, Labadia M, Grozinger K, Skoog M, Wu JC, Shih C-K, Eckner K, Hattox S, Adams J, Rosenthal AS, Faanes R, Eckner RJ, Koup RA, Sutton JL. Inhibition of HIV-1 replication by a nonnucleoside reverse transcriptase inhibitor. Science. 1990;250:1411–1413. doi: 10.1126/science.1701568. [DOI] [PubMed] [Google Scholar]
- 11.Figueiredo A, Moore KL, Mak J, Sluis-Cremer N, de Bethune M-P, Tachedjian G. Potent nonnucleoside reverse transcriptase inhibitors target HIV-1 Gag-Pol. PLoS Pathog. 2006;2:1051–1059. doi: 10.1371/journal.ppat.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young SD, Britcher SF, Tran LO, Payne LS, Lumma WC, Lyle TA, Huff JR, Anderson PS, Olsen DB, Carroll SS, Pettibone DJ, O'Brien JA, Ball RG, Balani SK, Lin JH, Chen I-W, Schleif SA, Sardana VV, Long WJ, Byrnes VW, Emini EA. L-743,726 (DMP-266): A novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 1995;39:2602–2605. doi: 10.1128/aac.39.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachedjian G, Orlova M, Sarafianos SG, Arnold E, Goff SP. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7188–7193. doi: 10.1073/pnas.121055998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venezia CF, Howard KJ, Ignatov ME, Holladay LA, Barkley MD. Effects of efavirenz binding on the subunit equilibria of HIV-1 reverse transcriptase. Biochemistry. 2006;45:2779–2789. doi: 10.1021/bi051915z. [DOI] [PubMed] [Google Scholar]
- 15.Sluis-Cremer N, Arion D, Parniak MA. Destabilization of the HIV-1 reverse transcriptase dimer upon interaction with N-acyl hydrazone inhibitors. Mol. Pharmacol. 2002;62:398–405. doi: 10.1124/mol.62.2.398. [DOI] [PubMed] [Google Scholar]
- 16.Sluis-Cremer N, Dmitrienko GI, Balzarini J, Camarasa M-J, Parniak MA. Human immunodeficiency virus type 1 reverse transcriptase dimer destablilization by 1-{spiro[4′′-amino-2′′,2′′-dioxo-1′′,2′′-oxathiole-5′′,3′-[2′,5′-bis-O-(tert-butyldimethylsilyl)-β-d-ribofuranosyl]]}-3-ethylthymine. Biochemistry. 2000;39:1427–1433. doi: 10.1021/bi991682+. [DOI] [PubMed] [Google Scholar]
- 17.Le Grice SFJ, Cameron CE, Benkovic SJ. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 1995;262:130–144. doi: 10.1016/0076-6879(95)62015-x. [DOI] [PubMed] [Google Scholar]
- 18.Ignatov ME, Berdis AJ, Le Grice SFJ, Barkley MD. Attenuation of DNA replication by HIV-1 reverse transcriptase near the central termination sequence. Biochemistry. 2005;44:5346–5356. doi: 10.1021/bi048000p. [DOI] [PubMed] [Google Scholar]
- 19.Tachedjian G, Aronson H-E, de los Santos M, Seehra J, McCoy JM, Goff SP. Role of residues in the tryptophan repeat motif for HIV-1 reverse transcriptase dimerization. J. Mol. Biol. 2003;326:381–396. doi: 10.1016/s0022-2836(02)01433-x. [DOI] [PubMed] [Google Scholar]
- 20.Chen RF. Fluorescence quantum yields of tryptophan and tyrosine. Anal. Lett. 1967;1:35–42. [Google Scholar]
- 21.Braz VA, Howard KJ. Separation of protein oligomers by blue native gel electrophoresis. Anal. Biochem. 2009;388:170–172. doi: 10.1016/j.ab.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh M, Jacques PS, Rodgers DW, Ottman M, Darlix JL, Le Grice SFJ. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 23.Tachedjian G, Aronson H-EG, Goff SP. Analysis of mutations and suppressors affecting interactions between subunits of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6334–6339. doi: 10.1073/pnas.97.12.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callis PR, Petrenko A, Muino PL, Tusell JR. Ab initio prediction of tryptophan fluorescence quenching by protein electric field enabled electron transfer. J. Phys. Chem. B. 2007;111:10335–10339. doi: 10.1021/jp0744883. [DOI] [PubMed] [Google Scholar]
- 26.Divita G, Restle T, Goody RS. Characterization of the dimerization process of HIV-1 reverse transcriptase heterodimer using intrinsic protein fluorescence. FEBS Lett. 1993;324:153–158. doi: 10.1016/0014-5793(93)81383-b. [DOI] [PubMed] [Google Scholar]
- 27.Barnard J, Borkow G, Parniak MA. The thiocarboxanilide nonnucleoside UC781 is a tight-binding inhibitor of HIV-1 reverse transcriptase. Biochemistry. 1997;36:7786–7792. doi: 10.1021/bi970140u. [DOI] [PubMed] [Google Scholar]
- 28.Venezia CF, Meany BJ, Braz VA, Barkley MD. Kinetics of association and dissociation of HIV-1 reverse transciptase subunits. Biochemistry. 2009;48:9084–9093. doi: 10.1021/bi9010495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Copeland RA. Evaluation of Enzyme Inhibitors in Drug Discovery. John Wiley & Sons; New Jersey: 2005. [Google Scholar]
- 30.Szedlacsek SE, Duggleby RG. Kinetics of slow and tight-binding inhibitors. Methods Enzymol. 1995;249:144–180. doi: 10.1016/0076-6879(95)49034-5. [DOI] [PubMed] [Google Scholar]
- 31.Sluis-Cremer N, Tachedjian G. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res. 2008;134:147–156. doi: 10.1016/j.virusres.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grobler JA, Dornadula G, Rice MR, Simcoe AL, Hazuda DJ, Miller MD. HIV-1 reverse transcriptase plus-strand initiation exhibits preferential sensitivity to non-nucleoside reverse transcriptase inhibitors in vitro. J. Biol. Chem. 2007;282:8005–8010. doi: 10.1074/jbc.M608274200. [DOI] [PubMed] [Google Scholar]
- 33.Lindberg J, Sigurdsson S, Löwgren S, Andersson HO, Sahlberg C, Noréen R, Fridborg K, Zhang H, Unge T. Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant. Eur. J. Biochem. 2002;269:1670–1677. doi: 10.1046/j.1432-1327.2002.02811.x. [DOI] [PubMed] [Google Scholar]
- 34.Xia Q, Radzio J, Anderson KS, Sluis-Cremer N. Probing nonnucleoside inhibitor-induced active site distortion in HIV-1 reverse transcriptase by transient kinetics analysis. Protein Sci. 2007;16:1728–1737. doi: 10.1110/ps.072829007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maga G, Ubiali D, Salvetti R, Pregnolato M, Spadari S. Selective interaction of the human immunodeficiency virus type 1 reverse transcriptase nonnucleoside inhibitor efavirenz and its thio-substituted analog with different enzyme-substrate complexes. Antimicrob. Agents Chemother. 2000;44:1186–1194. doi: 10.1128/aac.44.5.1186-1194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geitmann M, Unge T, Danielson H. Interaction kinetic characterization of HIV-1 reverse transcriptase non-nucleoside inhibitor resistance. J. Med. Chem. 2006;49:2375–2387. doi: 10.1021/jm0504050. [DOI] [PubMed] [Google Scholar]
- 37.Lohse A, Hardlei T, Jensen A, Plesner IW, Bols M. Investigation of the slow inhibition of almond β-glucosidase and yeast isomaltase by 1-azasugar inhibitors: evidence for the ‘direct binding’ model. Biochem. J. 2000;349:211–215. doi: 10.1042/0264-6021:3490211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narjes F, Brunetti M, Colarusso S, Gerlach B, Koch U, Biasiol G, Fattori D, De Francesco R, Matassa VG, Steinkühler C. α-Ketoacids are potent slow binding inhibitors of the hepatitis C virus NS3 protease. Biochemistry. 2000;39:1849–1861. doi: 10.1021/bi9924260. [DOI] [PubMed] [Google Scholar]
- 39.Borkow G, Fletcher RS, Barnard J, Arion D, Motakis D, Dmitrienko GI, Parniak MA. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-napthaldehyde hydrazone. Biochemistry. 1997;36:3179–3185. doi: 10.1021/bi9624696. [DOI] [PubMed] [Google Scholar]
- 40.Spence RA, Kati WM, Anderson KS, Johnson KA. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang D-E, Rizzo RC, Tirado-Rives J, Jorgensen WL. Antiviral drug design: computational analyses of the effects of the L100I mutation for HIV-RT on the binding of NNRTIs. Bioorg. Med. Chem. Lett. 2001;11:2799–2802. doi: 10.1016/s0960-894x(01)00510-8. [DOI] [PubMed] [Google Scholar]
- 42.Weikl TR, von Deuster C. Selected-fit versus induced-fit protein binding: kinetic differences and mutational analysis. Proteins. 2009;75:104–110. doi: 10.1002/prot.22223. [DOI] [PubMed] [Google Scholar]
- 43.Pegram LM, Record MT., Jr. Thermodynamic origin of Hofmeister ion effects. J. Phys. Chem. B. 2008;112:9428–9436. doi: 10.1021/jp800816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rösgen J, Pettitt BM, Bolen DW. Protein folding, stability, and solvation structure in osmolyte solutions. Biophys. J. 2005;89:2988–2997. doi: 10.1529/biophysj.105.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsiou Y, Ding J, Das K, Clark AD, Jr., Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanism. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 46.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 47.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 48.Willard HH, Merritt LL, Jr., Dean JA. Instrumental Methods of Analysis. Vol. 256. D. Van Nostrand Company; New Jersey: 1965. [Google Scholar]
- 49.Draper NR, Smith H. Applied Regression Analysis. Wiley & Sons; New York: 1966. pp. 77–81. [Google Scholar]
- 50.Efron B, Tibshrirani R. Bootstrap methods for standards errors, confidence intervals, and other measures of statistical accuracy. Stat. Sci. 1986;1:54–77. [Google Scholar]
- 51.Hall P. The Bootstrap and Edgeworth Expansion. Springer-Verlag; NY: 1992. p. 293. [Google Scholar]