Figure 3.
Detection of Alternative Splicing and 3D Protein Model
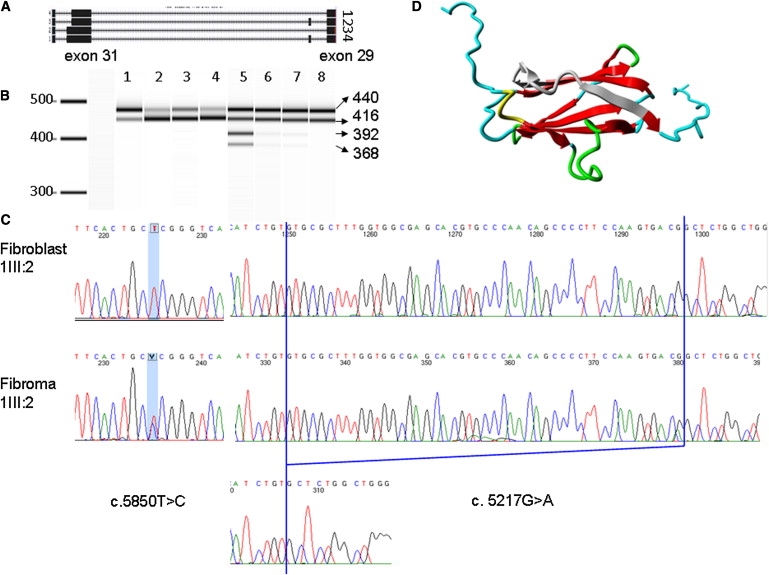
(A) Diagram of four FLNA transcripts in fibroma cells: transcripts 1 and 2, which carry the 48 bp deletion at the end of exon 31, as well as the normal transcripts 3 and 4.
(B) RT-PCR result from Agilent 2100 Bioanalyzer. Lane 1 is the product of the fibroblasts of 1III:6, which has a predominant longer isoform. Lanes 2–4 and 8 are four control human fibroblasts. Lanes 5–7 show RT-PCR products that were obtained from fibroma cells of 1III:6, the normal bands from two FLNA isoforms, and two extra shorter bands, which are faint in lane 6 (left fifth finger) and lane 7 (fifth toe of the left foot), whereas lane 5 (right fifth finger) shows four dark bands.
(C) Sanger sequencing results of c.5858T>C and c.5217G>A in fibroblast and fibroma cells of 1III:6.
(D) The 3D model of FLNA domain 15. The deleted 16 amino acids are marked in gray. Beta strands are marked in red. Green represents a turn. Yellow indicates a 3/10 helix. Random coils are colored in cyan.
