Figure 8.
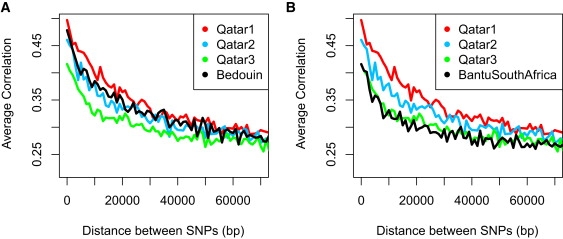
Linkage Disequilibrium Decay across the Genomes of the Qatari Subgroups and Two HGDP Population Samples
(A) Linkage disequilibrium (LD) for pairs of SNPs less than 70 kb apart was calculated as the squared correlation coefficient (r2). Calculations were performed on a standard sample size (n = 5) of randomly selected individuals in each Qatari group. SNP pairs were partitioned into bins in 1 kb intervals, and for each bin the mean r2 was plotted. The Qatar1 group has the highest LD, consistent with their higher degree of consanguinity. Qatar2 is intermediate, and the Qatar3 group has the lowest LD between SNPs, consistent with a large African component in their genome. The Bedouin HGDP population sample appears to fall between that of the Qatar1 and Qatar2 groups.
(B) The decay of LD of the three Qatari samples is replotted here along with the Bantu South African sample of the HGDP set. The LD decay of the Bantu South African population sample overlaps with that of Qatar3, consistent with the Qatar3 sample being of largely African origin.
