Abstract
The presence of infectious pancreatic necrosis virus (IPNV) in salmonids predominantly produces a high mortality rate in first-feeding fry. Genomic analysis of the vp2 gene sequence is most commonly used to determine the genetic diversity of IPNV isolates. Recently, information obtained from the vp1 gene allowed for efficient analysis of the genetic diversity of IPNV. In this study, the vp1 gene from a Mexican IPNV isolate was characterized and compared with IPNV isolates from Europe, North America, and Asia. The results indicate that the Mexican isolate is most closely related genetically to the 2310 strain from Spain.
Résumé
La présence du virus de la nécrose pancréatique infectieuse (IPNV) chez les salmonidés entraîne de façon prédominante un taux de mortalité élevé chez les alevins. L’analyse génomique de la séquence du gène vp2 est la plus communément utilisée pour déterminer la diversité génétique des isolats d’IPVN. Récemment, des informations obtenues à partir du gène vp1 ont permis une analyse efficace de la diversité génétique d’IPVN. Dans la présente étude, le gène vp1 provenant d’un isolat mexicain d’IPVN a été caractérisé et comparé à des isolats d’IPVN provenant d’Europe, d’Amérique du Nord et d’Asie. Les résultats indiquent que, génétiquement, l’isolat provenant du Mexique se rapproche le plus de la souche 2310 provenant d’Espagne.
(Traduit par Docteur Serge Messier)
In farm-raised rainbow trout (Oncorhynchus mykiss), infectious pancreatic necrosis virus (IPNV) has an important impact on population numbers owing to a high mortality rate among first-feeding fry. The fish that survive the infection become asymptomatic carriers and serve as a major means for the spread of disease.
The IPNV is a member of the family Birnaviridae, genus Aquabirnavirus. It has a double-stranded RNA (dsRNA) genome organized into 2 segments (A and B) within a nonenveloped, icosahedral capsid about 60 nm in diameter (1). The nucleotide sequence of segment A is approximately 3097 base pairs (bp) long and contains 2 open reading frames (ORFs) (2). The 1st ORF encodes the 17-kDa VP5 protein, which has only been found in infected cells (3). The 2nd ORF encodes a polyprotein precursor, known as NH2-pVP2-NS/VP4-VP3-COOH, which is cleaved by VP4 (also known as NS) to give rise to pVP2, VP3, and the same VP4 (4). The 54-kDa VP2 protein forms the capsid, the 31-kDa VP3 protein remains inside the virion, and VP4 is a protease involved in maturation of the virus. Segment B is approximately 2783 bp long and encodes the VP1 protein, which is found in 2 forms: a free polypeptide, RNA-dependent RNA polymerase (RdRp) (5); and the protein VPg, which is linked to the 5′ end of both genome segments (6).
The IPNVs have been classified into 2 serogroups and 9 serotypes (7). Isolates obtained from freshwater and marine fish have been found to belong to serotype A1 in the United States, the 4 serotypes A6, A7, A8, and A9 in Canada, and the 4 serotypes A2, A3, A4, and A5 in Europe (8). Representative A1, A2, and A3 serotypes have also been isolated in Asia and South America (9). Through an analysis of the Aquabirnavirus genome, a classification system has been proposed (10,11). In 2001 Blake et al (10) proposed 6 genogroups, defined by the amino acid sequence of the IPNV vp2 gene segment: genogroup I, the US isolates (A1) and 2 isolates from Canada (A9); genogroup II, isolates from Asia and Europe (A3); genogroup III, 2 isolates from Canada (C1 and ASV) (A6) and the European Te (A5); genogroup IV, isolates C2 (A7) and C3 (A8) from Canada; genogroup V, 5 European isolates and 1 isolate from Asia (A2); and genogroup VI, the isolate He (A4). In addition, Nishizawa et al (11) proposed in 2005 a genogroup VII, consisting of the isolates found in Japan and Korea.
In 2002 in Mexico an IPNV isolate from eyed eggs of rainbow trout from the United States was classified within serotype A1 and included in genogroup I according to vp2 sequence analysis (12). Several reports on the genetic diversity of IPNV have used the sequence of vp2, likely because this gene encodes the largest capsid protein, which is a more immunogenic protein, and is the most variable gene in IPNV (13). Although vp1 has also been considered for studying the genetic diversity of aquabirnaviruses (14), data on the diversity of this IPNV gene are currently limited. The vp1 gene encodes for the virus polymerase, and such genes are now widely recommended for RNA phylogenetic analysis (15). In this study, the sequence of vp1 of the Mexican isolate of IPNV was characterized and compared with that of several IPNV isolates from Europe, North America, and Asia, as well as 2 marine birnavirus (MABV) isolates.
A volume of 250 μL of CHSE-214 (Chinook salmon embryo cell line) cells cultured in Minimum Essential Medium supplemented with 10% bovine fetal serum infected with the Mexican IPNV isolate from rainbow trout (AF537289) was used to isolate total RNA with Trizol (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s instructions. The RNA was resuspended in 10 μL of water treated with 1% diethylpyrocarbonate (DEPC), and then 5 μL of the suspension was used for cDNA synthesis. The complete segment B was obtained by amplification of overlapping segments, with use of the primers described by Yao and Vakharia (16), with modifications (Table I).
Table I.
Characteristics of the primers used to amplify segment B of the cDNA genome of Mexican isolate AF537289 of infectious pancreatic necrosis virus (IPNV)
| Primera | Name | Nucleotide position | Product length (bp) | PCR Annealing temperature (°C) |
|---|---|---|---|---|
| 5′-GGAAACAGTGGGTCAACG-3′ | B-B′5NC | 1–18 | 500 | 55 |
| 5′-GAAGGTGAGTTGCTTCAGAAGTG-3′ | B-HindRa | 477–499 | ||
| 5′-GGAAGACGGCAAGCTTAAGGACAC-3′ | B-HindFa | 352–375 | 938 | 55 |
| 5′-GTGTTGTCCTGCAGTATGTAGATG-3′ | B-PstR | 1267–1290 | ||
| 5′-AGAGACAGCCTGGACAA-3′ | B-PstFa | 1217–1233 | 1072 | 51 |
| 5′-GAGTTTGGTCCTCTGGTCTAG-3′ | B-BstRa | 2285–2305 | ||
| 5′-AGAAAACCCGGAGCCGAGATTG-3′ | B-SmaΔFa | 1898–1919 | 886 | 60 |
| 5′-GGGGTCCCTGGCGGAACCGGATGT-3′ | B-Sma3′NC | 2761–2784 |
bp — base pairs; PCR — polymerase chain reaction.
Modified primers; bold type indicates nucleotides changed from those in the primers described by Yao and Vakharia (16).
Reverse-transcription polymerase chain reaction (RT-PCR) was performed in different tubes with use of the RevertAid M-MuLV Reverse Transcriptase Kit (Fermentas Canada, Burlington, Ontario). According to the manufacturer’s instructions, 1 μL (250 ng/μL) of the reverse primers (B-HindR, B-PstR, B-BstR, and B-Sma3′NC), 5 μL (1 μg/μL) of RNA, and 5 μL of DEPC water were mixed, and the mixture was incubated at 70°C for 10 min and then placed on ice. Next, 4 μL of 5X buffer (250 mM of Tris-HCl, pH 8.3, 250 mM of KCl, 20 mM of MgCl2, and 50 mM of DTT), 2 μL (4 mM mix) of deoxynucleotide triphosphates (dNTPs), 1 μL of RNAseOUT (20 U/μL), and 12 μL of DEPC water were added. The tubes were then incubated at 37°C for 5 min and placed on ice to add 40 units of the RT M-MuLV enzyme. Subsequently the tubes were incubated at 42°C for 60 min and then at 70°C for 10 min. The cDNA was stored at − 20°C until used.
The PCR was performed in different tubes for each primer pair with the use of 2.5 μL of cDNA, 5 μL of 10X ammonium reaction buffer (750 mM Tris-HCl, pH 8.5, 200 mM (NH4)2SO4, 15 mM MgCl2, and 1% Tween 20), 2.25 mM of MgCl2, 10 mM of dNTPs, 10 mM of each primer (B-B′5NC and B-HindR; B-HindF and B-PstR; B-PstF and B-BstR; B-SmaΔF and B-Sma3′NC), and 2.5 U of Taq polymerase (Gene Choice, San Diego, California, USA). The volume was brought up to 50 μL with DEPC-treated water. The reactions were performed in a thermocycler with the following conditions: initial denaturation at 94°C for 10 min, 35 cycles of denaturation at 94°C for 45 s, annealing temperature of each primer pair (Table I) for 45 s, elongation at 72°C for 60 s, and final elongation at 72°C for 10 min. The PCR products were subjected to electrophoresis in agarose gels stained with 2% ethidium bromide (0.5 μg/mL) at 100 V for 30 min and viewed with an ultraviolet transilluminator.
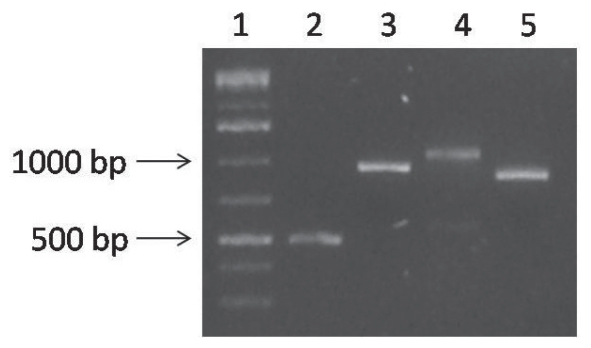
Fragments that matched the expected size of each reaction (Figure 1) were gel-purified (Wizard PCR and Gel Purification System; Promega, Madison, Wisconsin, USA) and cloned with use of the PCR II TOPO vector and a TOPO TA cloning kit (Invitrogen) according to the manufacturer’s instructions. The plasmid purifications were performed with the Wizard Plus SV Minipreps Kit (Promega), according to the manufacturer’s instructions. Subsequently, 2 mg of plasmid was used to obtain the forward and reverse strands of sequence nucleotides by means of a commercial kit (BigDye Terminator v. 3.1 Cycle Sequencing Kit; Applied Biosystems, Norwalk, Connecticut, USA) according to the manufacturer’s instructions. This assay was done in duplicate to obtain the complete nucleotide sequence of vp1 from the Mexican isolate, and the sequence was deposited in GenBank with the accession number EU665685. This sequence was compared with the 22 available complete sequences of IPNV vp1 in GenBank; their accession numbers and characteristics are presented in Figure 2B.
Figure 1.
Results of agarose gel electrophoresis of the products of polymerase chain reactions performed to obtain the complete segment B of the RNA genome of infectious pancreatic necrosis virus (IPNV). Column 1 — molecular marker; column 2 — product 500 base pairs (bp) long of B-B′5NC and B-HindR; column 3 — product 938 bp long of B-HindF and B-PstR; column 4 — product 1072 bp long of B-PstF and B-BstR; column 5 — product 886 bp long of B-SmaΔF and B-Sma3′NC.
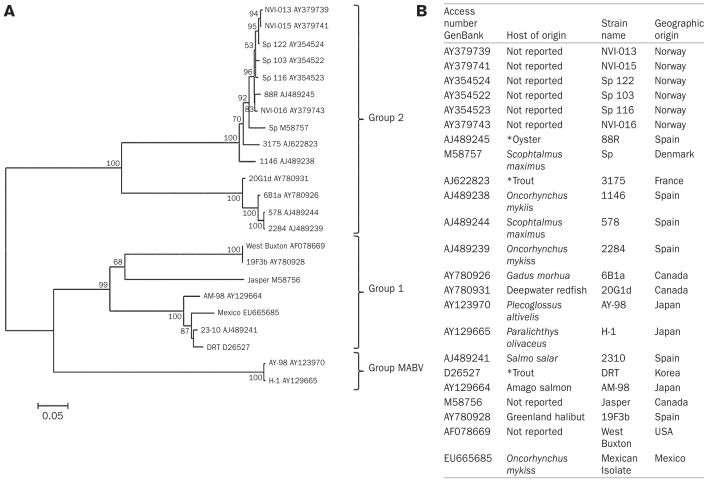
Figure 2.
A. Neighbor-Joining phylogenetic tree based on VP1 gene. Confidence on tree construction was assessed using 1000 bootstrap replicates (the values < 50% were not shown). The phylogenetic distance scale bar indicates estimated changes per nucleotide B. Table showing the characteristics of sequences obtained from the GenBank and its geographical origin used on phylogenetic tree.
*The scientific names of isolates host are not specific by authors.
Sequence alignments and molecular and phylogenetic analyses were performed with the use of MEGA 3.1 software (www.megasoftware.net). The sequences were analyzed by means of the Nei–Gojobori model with Jukes–Cantor modification (17). The phylogenetic tree was constructed by the neighbor-joining method and 1000 bootstrap replicates.
The nucleotide sequence of segment B of the Mexican isolate was found to be 2782 bp long. It begins with the conserved sequence GGAAA and ends with the sequence CCCC. The ORF consists of 2535 nucleotides, beginning with nucleotide 100 and ending with a TGA termination codon at nucleotide 2635. This ORF encodes a protein of 845 amino acids. Within the amino acid sequence the following motifs were found: 6 N-linked glycosylation sites (NXS/T), in the positions 184, 226, 409, 437, 658, and 677; 7 serine phosphorylation sites, in the positions 13, 21, 236, 245, 375, 635, and 701; 1 tyrosine phosphorylation site, in the position 399; and GLPYIGKT, DLEKGE, SGNAFTFLNN, LKN, and R, in the residues 248–255, 402–407, 471–480, 521–523, and 569, respectively.
For the phylogenetic analysis, we used a consensus sequence of 2535 nucleotides to compare our sequence with the 22 available in GenBank. The results showed a modified Nei–Gojobori genetic distance of 5.38 between the Mexican isolate and the 2310 strain and 97.75 between the Mexican isolate and the 578 and 2284 strains; all 3 comparison strains were from Spain. The distances between the Mexican isolate and the Jasper (Canada), Sp (Denmark), and West Buxton (United States) strains were 48.48, 91.43, and 41.62, respectively. An analysis of the nonsynonymous sequences demonstrated the modified Nei–Gojobori genetic distance between the Mexican isolate and the 2310 strain to be 0.61; the greatest distance was with the Sp (Denmark) strain, at 8.2.
The phylogenetic tree was organized into 3 groups: group 1, 7 isolates from Spain, Korea, Japan, Canada, United States, and the Mexican isolate; group 2, 14 isolates with origins in Spain, Norway, Denmark, France, and Canada. The group of MABV was formed for two MABV isolates originating in Japan (Figure 2A). The IPNV genomic segment that has been most widely used for studies of genetic diversity is the vp2 gene, presumably because the protein encoded by this gene plays an important role in the immune response and in the pathogenesis of the viral infection (13). Recently, studies of other birnaviruses have suggested that variations in RdRp could be related to changes in virulence and host specificity (18). Additionally, it has been reported that the vp1 nucleotide sequence is more conserved than the vp2 sequence; thus, sequence analysis of vp1 may allow for more precise phylogenetic studies. This was the basis for our interest in analyzing the sequence of vp1 and using this sequence for phylogenetic studies.
We found that the nucleotide sequence of segment B of the Mexican IPNV isolate consists of 2782 bp, unlike the Sp (Denmark) strain (2630 bp) but similar to the Jasper strain (2784 bp) and the West Buxton strain (2783 bp). Compared with the sequence of the Jasper strain, that of the Mexican isolate has 2 deletions, 1 in the 5′ noncoding region and the other in the 3′ noncoding region. The ORF of vp1 encodes a protein of 844 amino acids in the Sp (Denmark) strain, whereas in the Jasper and West Buxton strains (16,19) and in the Mexican isolate it encodes a protein of 845 amino acids. According to BLAST analysis, the highest percentage of identity in the amino acid sequence of the Mexican isolate is 98% with the 2310 isolate (CAD32983.1 in GenBank), which belongs to genogroup I, as does the Mexican isolate.
Several motifs have been found in the IPNV RdRp, 1 of which is GXXXXGKS/T (where X represents any amino acid), a conserved motif in guanosine triphosphate-binding proteins. This motif has been observed in several viral proteins that have a putative role in the replication of RNA (20) and was also found in the Mexican IPNV isolate, between residues 248 and 255 (GLPYIGKT). The GDD sequence is a conserved motif that is present in the RdRp of almost all RNA viruses, but this motif is substituted with the motif LKN or LKD in IPNV (19). We determined that the Mexican isolate contains the motif LKN in residues 521 and 523. Interestingly, this motif has also been reported in other aquabirnaviruses (19). The motifs DXXXXE and SGXXXTXXXN have also been identified in IPNV and marine birnaviruses (19,21); in the Mexican IPNV isolate they correspond to the residues 402 to 407 (DLEKGE) and 471 to 480 (SGNAFTFLNN). A motif was also found in the Mexican IPNV isolate with a single residue (R) at position 569; although this motif has not been reported in other IPNV isolates, it has been reported in infectious bursal disease virus (22). Furthermore, we found 6 N-linked glycosylation sites, 7 serine phosphorylation sites, and 1 tyrosine phosphorylation site that had previously been reported (14); however, we did not find the serine phosphorylation site at position 802.
In the phylogenetic tree analysis, we observed 3 groups: group 1, formed by 2 subgroups (West Buxton, 19F3b, and Jasper; and AM-98, 2310, DRT, and the Mexican isolate); group 2, formed by the Canadian strains 6B1a and 20G1d, the Sp strains, and other isolates from Europe; and a group that contained only the marine birnavirus isolates AY-98 and H1. Interestingly, the sequences of isolates 6B1a and 20G1d showed 3 insertions, at the 770, 782, and 799 positions, and the modified Nei–Gojobori genetic distance between them was 3.95, whereas the distances between these isolates and the Sp (Denmark) strain were 55.07 and 48.74, respectively.
Our data coincided with reports published by Nishizawa et al (11) and Blake et al (10), both of which described a group containing different Sp strains with other isolates and another group containing the West Buxton and Jasper strains along with others, including the Mexican isolate. Recently, a study using sequences from the vp2 of other IPNV isolates obtained in Mexico showed that the West Buxton and Jasper strains are genetically related to the Mexican isolate AF537289 (23), which is consistent with the results we obtained using vp1. On the other hand, our analysis also indicates that marine birnaviruses form a group that is separate from IPNV, in agreement with the work of Zhang and Suzuki (24). Although the Mexican isolate is in the same group as the West Buxton and Jasper strains according to the phylogenetic tree, the similarity of the vp1 genes indicates that the Mexican isolate is more closely related genetically to strains 2310 from Spain, DRT from Korea, and AM-98 from Japan, with modified Nei–Gojobori genetic distances of 5.38, 6.35, and 9.48, respectively.
Global information about vp1 nucleotide sequences is scarce. In this work we characterized the vp1 of a Mexican IPNV isolate. Since this gene is more conserved than vp2, we conclude that phylogenetic analysis of vp1 is more suitable for identifying new genetic lineages in future studies.
Acknowledgments
We thank Alicia López and Abel Sánchez for their technical assistance. The authors are especially grateful to the Universidad Autónoma del Estado de México. This work was supported by Consejo Nacional de Ciencia y Tecnología.
References
- 1.Dobos P, Hill BJ, Hallett R, Kells DTC, Becht H, Teninges D. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J Virol. 1979;32:593–605. doi: 10.1128/jvi.32.2.593-605.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan R, Dobos P. The nucleotide sequence of infectious pancreatic necrosis virus (IPNV) dsRNA segment A reveals one large ORF encoding a precursor polyprotein. Nucleic Acids Res. 1986;14:5934. doi: 10.1093/nar/14.14.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magyar G, Dobos P. Evidence for the detection of the infectious pancreatic necrosis virus polyprotein and the 17-kDa polypeptide in infected cells and of the NS protease in purified virions. Virology. 1994;204:580–589. doi: 10.1006/viro.1994.1572. [DOI] [PubMed] [Google Scholar]
- 4.Petit S, Lejal N, Huet JC, Delmas B. Active residues and viral substrate cleavage sites of the protease of the birnavirus infectious pancreatic necrosis virus. J Virol. 2000;74:2057–2066. doi: 10.1128/jvi.74.5.2057-2066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobos P. Protein-primed RNA synthesis in vitro by the virion associated RNA polymerase of infectious pancreatic necrosis virus. Virology. 1995;208:19–25. doi: 10.1006/viro.1995.1125. [DOI] [PubMed] [Google Scholar]
- 6.Calvert JG, Nagy E, Soler M, Dobos P. Characterization of the VPg-dsRNA linkage of infectious pancreatic necrosis virus. J Gen Virol. 1991;72:2563–2567. doi: 10.1099/0022-1317-72-10-2563. [DOI] [PubMed] [Google Scholar]
- 7.Hill BJ, Way K. Serological classification of infectious pancreatic necrosis (IPN) virus and other aquatic birnaviruses. Annu Rev Fish Dis. 1995;5:55–77. [Google Scholar]
- 8.Romero-Brey I, Batts WN, Bandín I, Winton JR, Dopazo CP. Molecular characterization of birnaviruses isolated from wild marine fishes at the Flemish Cap (Newfoundland) Dis Aquat Organ. 2004;61:1–10. doi: 10.3354/dao061001. [DOI] [PubMed] [Google Scholar]
- 9.Reno PW. Infectious pancreatic necrosis and associated aquatic birnaviruses. In: Woo PTK, Bruno DW, editors. Fish Diseases and Disorders Vol 3: Viral, Bacterial and Fungal Infections. London, England: CABI Publishing; 1999. pp. 3–55. [Google Scholar]
- 10.Blake S, Ma JY, Caporale DA, Jairath S, Nicholson BL. Phylogenetic relationships of aquatic birnaviruses based on deduced amino acid sequences of genome segment A cDNA. Dis Aquat Organ. 2001;45:89–102. doi: 10.3354/dao045089. [DOI] [PubMed] [Google Scholar]
- 11.Nishizawa T, Kinoshita S, Yoshimizu M. An approach for genogrouping of Japanese isolates of aquabirnaviruses in a new genogroup, VII, based on the VP2/NS junction region. J Gen Virol. 2005;86:1973–1978. doi: 10.1099/vir.0.80438-0. [DOI] [PubMed] [Google Scholar]
- 12.Ortega SC, Montes de Oca R, Groman D, Yason C, Nicholson B, Blake S. Case report: Viral infectious pancreatic necrosis in farmed rainbow trout from Mexico. J Aquat Anim Health. 2002;14:305–310. doi: 10.1577/1548-8667(2002)014<0305:CRVIPN>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Santi N, Vakharia VN, Evensen O. Identification of putative motifs involved in the virulence of infectious pancreatic necrosis virus. Virology. 2004;322:31–40. doi: 10.1016/j.virol.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Joh SJ, Shon CI, Kang SW, et al. Molecular characterization and genogrouping of VP1 of aquatic birnavirus GC1 isolated from rockfish Sebastes schlegeli in Korea. J Vet Sci. 2008;9:85–90. doi: 10.4142/jvs.2008.9.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kooning EV, Dolja VV. Evolution and taxonomy of positive-strand viruses: implications and analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 16.Yao K, Vakharia VN. Generation of infectious pancreatic necrosis virus from cloned cDNA. J Virol. 1998;72:8913–8920. doi: 10.1128/jvi.72.11.8913-8920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 18.Liu M, Vakharia VN. VP1 protein of infectious bursal disease virus modulates the virulence in vivo. Virology. 2004;330:62–73. doi: 10.1016/j.virol.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Duncan R, Mason C, Nagy E, Leong J-A, Dobos P. Sequence analysis of infectious pancreatic necrosis genome segment B and its encoded VP1 protein: A putative RNA-dependent RNA polymerase lacking the Gly–Asp–Asp motif. Virology. 1991;181:541–552. doi: 10.1016/0042-6822(91)90887-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang CX, Suzuki S. Comparison of the RNA polymerase genes of marine birnavirus strains and other birnaviruses. Arch Virol. 2003;148:745–758. doi: 10.1007/s00705-002-0951-y. [DOI] [PubMed] [Google Scholar]
- 22.Islam MR, Zierenberg K, Muller H. The genome segment B encoding the RNA-dependent RNA polymerase protein VP1 of very virulent infectious bursal disease virus (IBDV) is phylogenetically distinct from that of all other IBDV strains. Arch Virol. 2001;146:2481–2492. doi: 10.1007/s007050170018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrero L, Herrera E, Salinas J, Torres J, Montero AB, Barrón B. Detection and genotyping of an infectious pancreatic necrosis virus from asymptomatic rainbow trout (Oncorhynchus mykiss) facilities in Mexico. Intervirology. 2008;51:285–292. doi: 10.1159/000170903. Epub 2008 Nov 6. [DOI] [PubMed] [Google Scholar]
- 24.Zhang CX, Suzuki S. Aquabirnaviruses isolated from marine organisms form a distinct genogroup from other aquabirnaviruses. J Fish Dis. 2004;27:633–643. doi: 10.1111/j.1365-2761.2004.00585.x. [DOI] [PubMed] [Google Scholar]