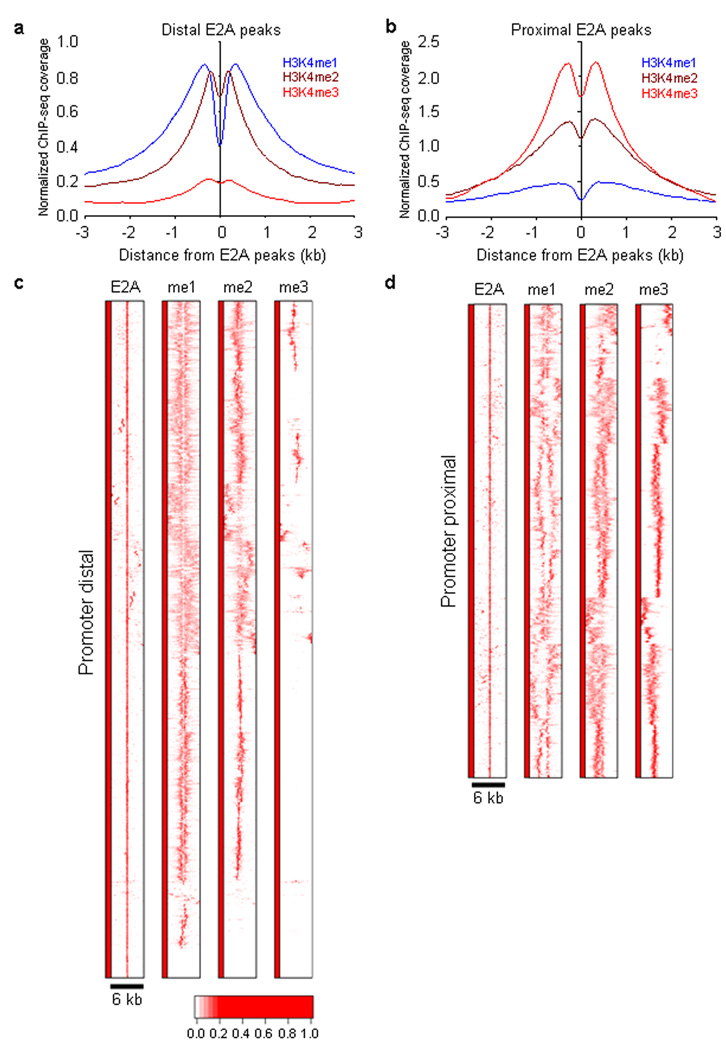
Figure 2.
E2A occupancy and H3K4 methylation in cultured RAG1-deficient pro-B cells. (a) Heat map of E2A occupancy and H3K4me1, H3K4me2 and H3K4me3 patterns for promoter-distal DNA sequences (> 3 kb separation from nearby transcription start sites). Each row represents a 6 kb genomic region showing E2A occupancy. The heat map reflects the enrichment of E2A binding and H3K4 methylation by color according a scale as indicated below. (b) Heat map of E2A occupancy and distributions of H3K4me1, H3K4me2 and H3K4me3 for promoter-proximal (< 3kb from nearby transcription initiation sites) region. (c) E2A occupancy correlates with H3K4 mono- and di-methylation in promoter distally located regions. The genomic distance distributions of H3K4 methylation (H3K4me1, H3K4me2 and H3K4me3) centered across E2A occupancy are shown across a 6 kb genomic region. (d) E2A occupancy correlates with H3K4 mono- and di-methylation in promoter-proximal located regions. The genomic distance distributions of H3K4 methylation (H3K4me1, H3K4me2 and H3K4me3) centered across E2A occupancy are shown across a 6 kb genomic region. A total of four independent ChIP-Seq experiments were performed in order to determine genome-wide E2A occupancy as well as patterns of H3K4me1, H3K4me2 and H3K4me3.