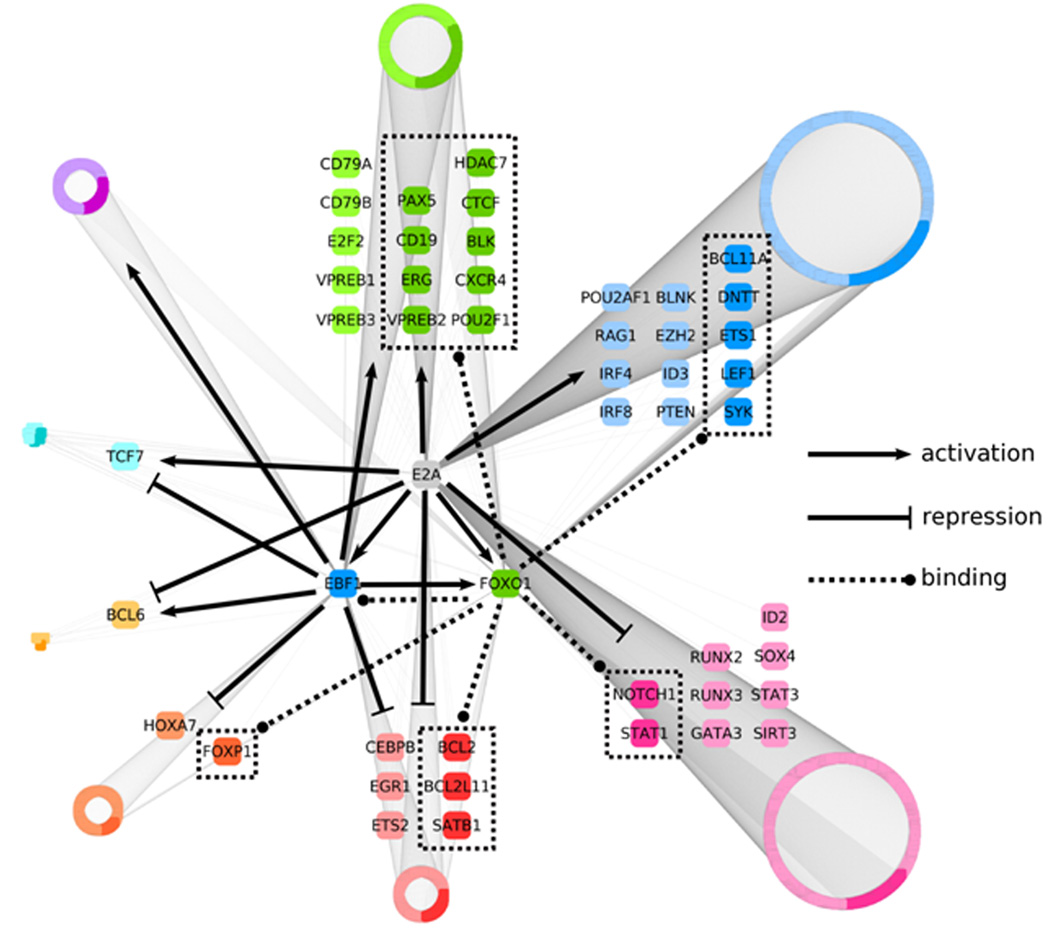
Figure 8.
Regulatory network that links the activities of an ensemble of transcriptional regulators, signaling components and survival factors in developing B cells into a common pathway. An integrative genome-wide analysis, using Cytoscape software, of protein-DNA binding and changes in transcript levels across E2A-deficient pre-pro-B, EBF1-deficient pre-pro-B and pro-B cells was used to identify transcriptional regulatory targets of E2A and EBF1, as well as genes bound by FOXO1. Resulting 2045 regulatory targets were divided into eight groups based on positive or negative changes in transcript levels in pro-B cells as compared to E2A or EBF1-deficient cells. Arrows represent gene activation whereas barred lines indicate transcriptional repression. Dotted lines represent FOXO1 occupancy. Selected genes from each group either known to play critical roles in B cell commitment or potential novel regulators are enlarged. All other genes are grouped into rings where the size of each ring indicates the relative number of members. Darker colors present loci that show FOXO1 occupancy.