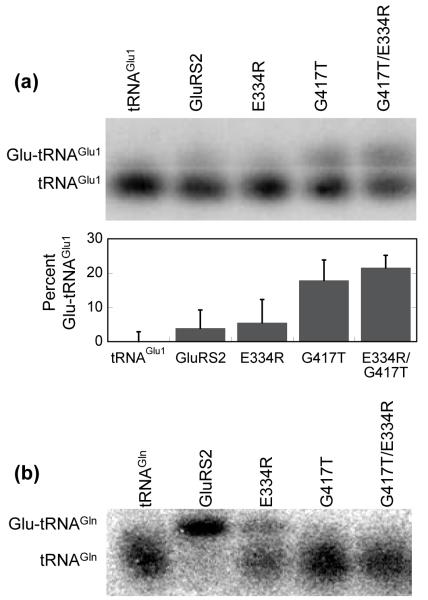
Figure 4.
Disruption of Gly417 in Hp GluRS2 leads to tRNAGlu aminoacylation and eliminates tRNAGln aminoacylation. (a) Aminoacylation of tRNAGlu1 by Gly417Thr and Glu334Arg/Gly417Thr GluRS2. Transfer RNAGlu1 was separately aminoacylated by wild-type GluRS2 (lane 2), Glu334Arg GluRS2 (lane 3), Gly417Thr GluRS2 (lane 4), or the double mutant Glu334Arg/Gly417Thr GluRS2 (lane 5), as described for Figure 2. Lane 1 shows deacylated tRNAGlu1 as a control. Acid gels and Northern blots were performed as described in Figure 2. Gel shown is representative of experiments run in triplicate. The percent Glu-tRNAGlu1 formed was determined using ImageQuant software (Molecular Dynamics) to quantify the ratio of the upper and lower bands. The bar graph shows the results of this analysis; Lane 1 (deacylated tRNAGlu1) was used as a background control and subtracted from lanes 2-5. (b) Aminoacylation of tRNAGln by Hp GluRS2 and various mutants. Transfer RNAGln was separately aminoacylated as in Figure 4a. Lane 1 shows deacylated tRNAGln as a control.