Abstract
Mitogen- and stress-activated protein kinase 1 (MSK1) is a growth factor-stimulated serine/threonine kinase that is involved in gene transcription regulation and proinflammatory cytokine stimulation. MSK1 is a dual kinase possessing two non-identical protein kinase domains in one polypeptide. We present the active conformation of the crystal structures of its C-terminal kinase domain (CTD) in apo form and complexed with a non-hydrolyzable ATP analogue at 2.0 Å and 2.5 Å resolution, respectively. Structural analysis revealed substantial differences in the contacts formed by the C-terminal helix, which is responsible for the inactivity of other autoinhibited kinases. In CTD MSK1, the C-terminal αL-helix is located in the surface groove, but forms no hydrogen bonding with the substrate-binding loop or nearby helices, and does not interfere with the protein's autophosphorylation activity. Mutational analysis confirmed that the αL-helix is inherently non-autoinhibitory. Overexpression of the single C-terminal kinase domain in JB6 cells resulted in tumor promoter-induced neoplastic transformation in a manner similar to that induced by the full length MSK1 protein. The overall results suggest that the CTD MSK1 is regulated by a novel, αL-helix-independent mechanism, suggesting that a diverse mechanism of autoinhibition and activation might be adopted by members of closely related protein kinase family.
Keywords: crystal structure, protein kinase, autoinhibitory helix, active form, autophosphorylation
Introduction
Mitogen- and stress-activated protein kinase 1 (MSK1) is a serine/threonine kinase that is activated by growth factors, mitogens, proinflammatory cytokines and stress. MSKs are involved in the regulation of transcription downstream of mitogen-activated protein (MAP) kinases. 1,2 They phosphorylate a number of proteins, including histone H3 3-5, cAMP-response-element-binding protein 6, activating transcription factor 7, nuclear factor-κB 8, transcription factor ER81 9 and chromatin-associated high-mobility group 14 proteins 10. MSKs are involved in the transcriptional induction of immediate-early genes c-fos and Nur77 11-13 and play an important role in stimulating the production of anti-inflammatory cytokines 14. MSKs are attractive targets for the treatment of chronic inflammatory diseases 15. MSK1 is required for tumor promoter-induced neoplastic cell transformation 16.
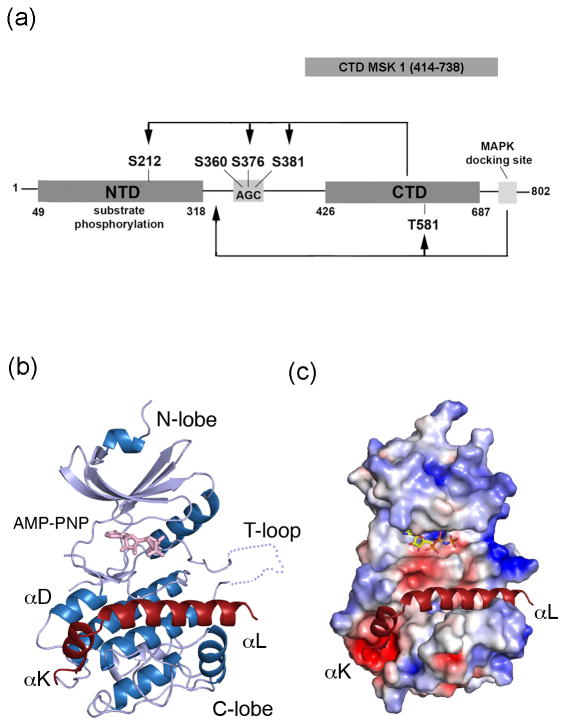
MSK1 belongs to the p90 ribosomal S6 kinase (RSK) protein family. Members of this family have a similar architecture – a single polypeptide with two distinct protein kinase domains connected by a hydrophobic linker region (Fig. 1a). The N-terminal kinase domain of MSK1 (NTD MSK1) is responsible for phosphorylation of a variety of substrates. The C-terminal kinase domain of MSK1 (CTD MSK1) is an important regulatory domain that is activated by extracellular signal-regulated kinases (ERKs) and p38 mitogen-activated protein (MAP) kinase in response to various cellular stimuli. The complete activation of full length MSK1 is controlled by multiple phosphorylation sites 17,18, which is initiated through the stimulation of its C-terminal kinase domain. The CTD MSK1 is activated by phosphorylation of Thr581 in its T-activation loop by ERKs/or p38. The activated CTD is believed to subsequently phosphorylate Ser376 and Ser381 in the hydrophobic linker region and Ser212 in the T-loop of the NTD MSK1 leading to complete activation of the full length MSK1 protein (Fig. 1a).
Figure 1. The kinase scaffold of the CTD MSK1.
(A) A schematic diagram of the full length MSK1. MSK1 possesses two distinct protein kinase domains that are sequentially activated by phosphorylation at multiple sites. The serine 360, 376, and 381 residues in the hydrophobic linker region are located within the conserved AGC-kinase sequence. The arrow indicates the sites phosphorylated by CTD MSK1 and MAP kinases. A ribbon diagram (B) and electrostatic surface (C) representation of CTD MSK1 (residues 414-738) are shown. The helices and strands are colored in blue and silver, respectively. The C-terminal extension (i.e, the αK and αL-helices) is shown in red, the AMP-PNP molecule is shown in sticks (pink), and the disordered T-loop is shown as a dotted line.
As indicated earlier, the C-terminal and N-terminal domains of MSK1 are structurally distinct kinases. The NTD MSK1 belongs to the AGC kinase family that is characterized by a kinase domain followed by a hydrophobic motif region with phosphorylation sites. The crystal structure of the NTD MSK1 is available 19, whereas the single CTD MSK1 and the full length MSK1 protein structures have not yet been solved. The crystal structure of the C-terminal kinase domain of the related p90 ribosomal S6 kinase 2 (CTD RSK2) was recently determined 20 and showed a scaffold similarity with the MAP kinase-activated protein kinase 2 (MK2). MK2 is a serine/threonine kinase that is activated by p38 MAP kinase in response to cellular stress 21-23. MK2 is exclusively activated by p38 and the CTD RSK2 is activated by ERK1/2. Notably, the CTD MSK1 is activated by either p38 or ERK1/2. Phylogenetically, the CTD MSK1 is related to MK2 and these proteins share 35% sequence homology 24,25. MSK1, a dual kinase domain protein, and MK2, a single kinase-domain protein, each have a MAP kinase docking site and a putative bipartite nuclear localization signal at the C-terminus. The MK2 structure has been reported in an inactive 26 and active conformation in complex with ADP 27 and inhibitors 28,29.
Our study describes the crystal structure of the isolated active CTD MSK1 (residues 414-738) in apo form and in complex with the non-hydrolyzable ATP analogue, AMP-PNP. We showed that, despite an overall similarity with RSK2, the CTD MSK1 is unexpectedly not autoinhibited by the C-terminal helix. Furthermore, the CTD MSK1 undergoes active autophosphorylation in vitro and induces neoplastic cell transformation ex vivo. The intrinsic non-autoinhibitory C-terminal αL-helix feature suggests that the CTD MSK1 is stimulated by an αL-helix-independent activation mechanism. In addition to structure elucidation, our data indicate that the extreme C-terminal end, which encompasses the MAP kinase-docking site, might regulate CTD MSK1 activity.
Results
Crystal structure of the CTD MSK1
The crystal structures of several CTD MSK1 fragments were determined and the best diffraction data were obtained for the construct comprising residues 414-738. The refined crystal structures included the CTD MSK (residues 415-729), in apo form and in complex with the non-hydrolyzable ATP analogue, AMP-PNP, determined at 2.0 Å and 2.5 Å resolution, respectively. The data collection and refinement statistics are presented in Table 1. Results indicated that the CTD MSK1 adopts a classical kinase bilobal scaffold with the AMP-PNP molecule bound to a glycine-rich loop (Fig. 1b). The C-terminal segment (residues 700-729) extends from the putative kinase domain and adopts a helix-turn-helix conformation. The short αK-helix (residues 700-705) and the longer αL-helix (residues 709-729) are located within deep grooves on the surface (Fig. 1b and 1c). The asymmetric unit contains two highly similar and well-ordered molecules (Supplementary Figure S1). Size exclusion chromatography indicated that, in solution, the protein used for crystallization was monomeric. The observed dimerization was likely driven by increasing protein concentration during crystallization.
Table 1. X-ray data collection and refinement statistics.
| Data Collection | MSK (apo-) | MSK/AMP-PNP |
|---|---|---|
| Wavelength (Å) | 0.979 | 0.979 |
| Space group | P212121 | P212121 |
| Unit cell (Å) | a=51.3 b=91.7, c=134.9 | a=51.5 b=91.2, c=135.4 |
| Resolution (Å) | 2.0 | 2.5 |
| Highest shell (Å) | 2.15-2.0 | 2.70-2.50 |
| Total observations | 306,829 (23,896) | 114,102 (8,154) |
| Unique reflections | 43,534 (4,120) | 23,120 (2,204) |
| I / σI | 31.4 (3.1) | 16.0 (2.8) |
| Rsym (%)† | 9.1 (44.1) | 11.8 (49.2) |
| Completeness (%) | 99.4 (99.5) | 96.3 (94.1) |
| Refinement | ||
| Resolution range (Å) | 45-2.0 | 43-2.5 |
| No. Reflections | 43,443 | 20,472 |
| Rfree/Rfactor (%)† | 26/21 | 27/20 |
| No. atoms: | ||
| Total | 4,744 | 4,704 |
| Protein | 4,535 | 4,620 |
| Solvent | 209 | 84 |
| Mean B values (Å2): | ||
| Protein | 27.8 | 53.4 |
| AMP-PNP | - | 68.9 |
| Bond lengths (Å) | 0.005 | 0.006 |
| Bond angles (deg) | 0.98 | 1.1 |
| Ramachandran most favored positions | 99% | 99% |
One crystal was used for the structure determination.
Highest resolution shell is shown in parenthesis.
Rfree was calculated from a randomly chosen 5% of reflections excluded from refinement
as defined in CCP4 38.
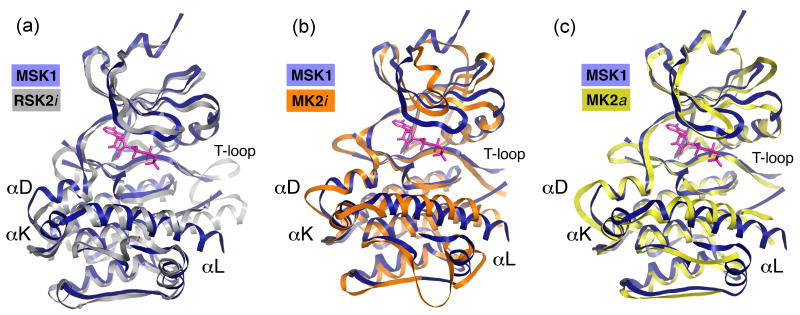
The overall kinase scaffold of the CTD MSK1 resembles that of the autoinhibited CTD RSK2 with an r.m.s.d. of 1.48 Å for the 250 corresponding Cα atoms (Fig. 2a). The most structurally divergent region between both proteins resides in the large α-helical C-lobe. Unlike the CTD RSK2, which has a coil, the CTD MSK1 possesses an additional short αK-helix immediately preceding the C-terminal αL-helix. The T-activation loop (residues 575-596) of CTD MSK1 is disordered and not visible in the electron density map. However, the loop is likely to be elongated compared to RSK2 because of the shorter αF-helix. The αF-helix of RSK2 is unusually long, resulting in a shortening of the T-loop turned away from the catalytic cleft.
Figure 2. A comparison of the structure of the CTD MSK1/AMP-PNP complex with the inactive CTD RSK2 and inactive/active MK2.
The CTD MSK1 structure is shown in blue. The AMP-PNP molecule in pink is depicted from the CTD MSK1 structure. (A) The inactive CTD RSK2 (PBD code 2QR8) is shown in silver. The two kinases display differences in the C-lobe. Note that the CTD MSK1 has a novel short αK-helix and a longer αL-helix. The T-activation loop of the CTD MSK1 is disordered. (B) The inactive MK2 (PDB code 1KWP) is shown in orange. The C-terminal coil of MK2 (residues 364-385) is not shown for clarity. MK2 and CTD MSK1 exhibit major differences in the N-lobe, especially in the orientation of the phosphate-binding loop and the bottom region of the C-lobe. In contrast to the corresponding MK2 helix, the C-terminal MSK1 αL-helix does not form hydrogen bonds with the kinase C-lobe. (C) The active MK2 (PDB code 2P3G) is shown in yellow. Among the available truncated MK2 structures in the active conformation, only this structure showed the presence of an intact C-terminal helix (residues 350-359).
We superimposed the CTD MSK1 structure with the structures of MAP kinase-activated protein kinase 2 (MK2) in the various available conformations (Fig. 2b and 2c). The comparison of the CTD MSK1 with inactive MK2 (MK2i) showed substantial differences especially in the phosphate-binding loop position. The CTD MSK1 structure superimposed on the active MK2 structure (MK2a) exhibits definite differences in the position of some secondary elements. However, similar to the active/inactive MK2, the C-terminal extension of the CTD MSK1 adopts a helix-turn-helix conformation with the αK- and αL-helices embedded in the kinase scaffold. We observed that the CTD MSK1 C-terminal αL-helix occupies the same position in the surface groove as the corresponding helix of the autoinhibited CTD RSK2 and both the inactive/active MK2.
The isolated CTD MSK1 is active in vitro
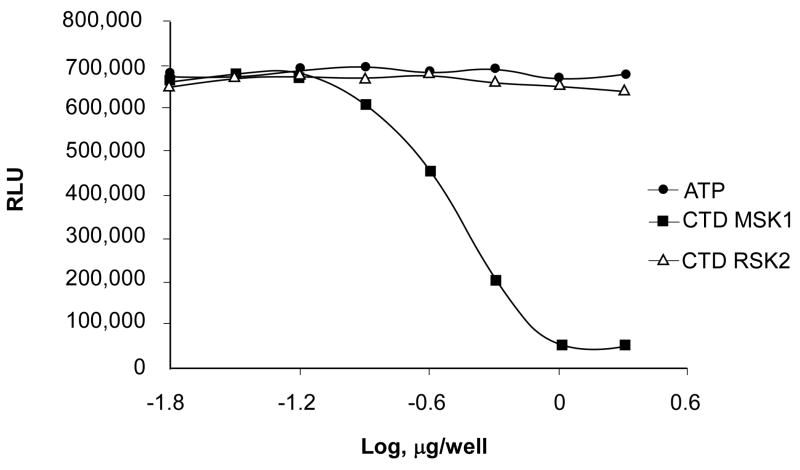
To determine whether the recombinant CTD MSK1 is active, we performed a kinase assay with [γ-32P] ATP and a customized peptide encompassing residues from the linker region as substrate. As indicated earlier, the CTD MSK1 is believed to phosphorylate two serine residues in the hydrophobic linker region and Ser212 in the T-activation loop of the N-terminal domain. Our results indicated that neither the commercially available active full length MSK1 nor the isolated CTD MSK1 could phosphorylate the peptide-linker region. However, the CTD MSK1 could autophosphorylate itself (data not shown). Therefore, to assess the autophosphorylation activity of the CTD MSK1, we performed a Kinase-Glo® Luminescent Assay. This assay measures kinase activity by quantifying the amount of ATP remaining in solution at the completion of the kinase reaction. The CTD MSK1 protein underwent active autophosphorylation as was measured by the decrease in Relative Luminescent Units (Fig. 3). In contrast, the inactive CTD RSK2 neither autophosphorylated itself in solution nor bound ATP in the crystals.
Figure 3. The kinase luminescent autophosphorylation assay.
CTD MSK1 autophosphorylation was assessed using a Kinase-Glo® Luminescent Assay as described in Experimental Procedures. The luminescent signal is represented as Relative Luminescence Units (RLU) and is inversely correlated with kinase activity. The CTD MSK1 (414-738 residues) autophosphorylation reaction was complete with as little as 1 μg protein/well. The CTD RSK2 (399-740 residues) did not show any autophosphorylation activity up to 20 μg protein/well. The experiment was repeated at least 3 times with purified proteins from separate preparations.
The C-terminal αL-helix of the CTD MSK1 is inherently non-autoinhibitory
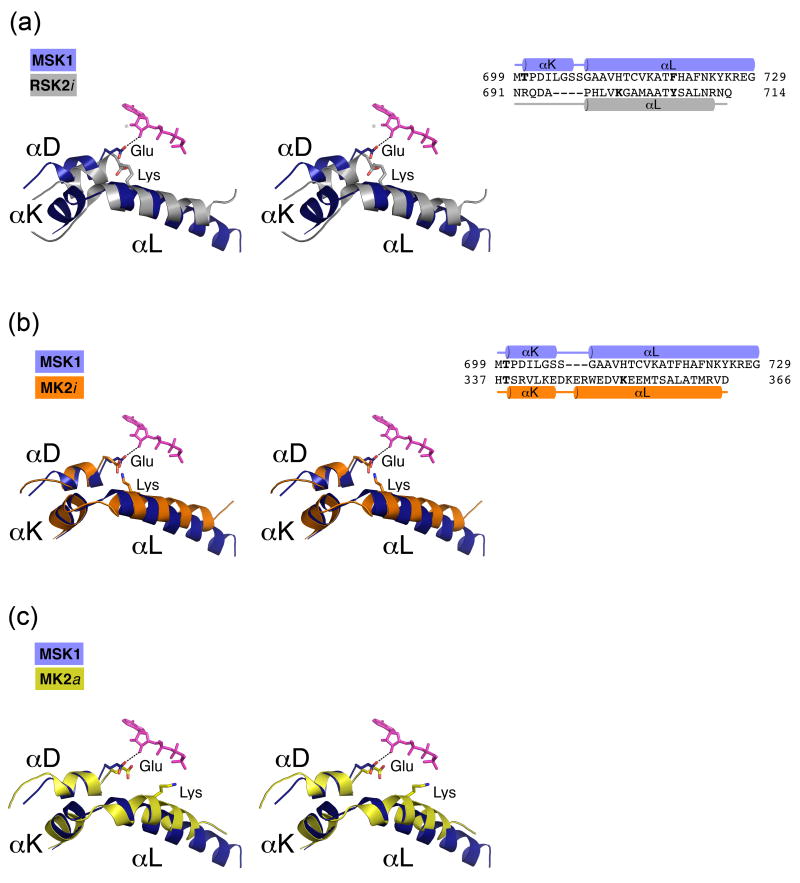
The C-terminal αL-helix of CTD MSK1 does not prevent ATP-binding in the crystals or protein autokinase activity as was shown above. In order to determine why the CTD MSK1 is active, we focused on the specific amino acid residues involved in the αL-helix interactions with the kinase domain in the CTD MSK1, and then compared them with the autoinhibited CTD RSK2 and MK2 and the active MK2. In the inactive CTD RSK2, the Tyr707 residue located on the C-terminal autoinhibitory αL-helix forms a single hydrogen bond with the Ser603 on the αF-helix, and disruption of that contact by mutation of Tyr707 to alanine resulted in the activation of RSK2 20,30,31. In the case of MSK1, the corresponding Tyr707RSK2 is replaced by Phe719, which cannot form a hydrogen bond. Only the hydrophobic side chain of the MSK1 residue, Phe722 that is located in the middle of the αL-helix, created a weak hydrophobic stacking interaction (3.2 Å) with the aromatic ring of Trp606 on the αF-helix. Another important CTD RSK2 residue, Lys700 that is located on the autoinhibitory αL-helix, attracted the Glu500 from the αD-helix and caused misalignment of the Glu500 residue, one of the conserved residues participating in ATP binding (Fig. 4a). The CTD RSK2 activation was hypothesized to occur through the αL-helix displacement in the surface groove, which triggers the αD-helix reposition and proper alignment of the Glu500 residue 20. Remarkably, the CTD MSK1 does not have a corresponding positively charged residue on the αL-helix, and lacks the homologous lysine-glutamate salt bridge (see sequence alignment in Fig. 4a). As a result, the Glu505MSK1 residue, one of the conserved residues participating in ATP binding, can form a weak hydrogen bond (3 Å) with the ATP ribose ring. The Oε1 and Oε2 atoms of Glu505MSK1 are shifted by 3.7 and 3.0 Å, respectively, compared to the corresponding atoms of the homologous Glu500RSK2. The entire αD-helix of the active CTD MSK1 is shifted by 1.3 Å compared with the inactive CTD RSK2 as was previously predicted for RSK2 activation.
Figure 4. An enlarged view of the αL/αD helix interaction (left) and the 3D-structure based alignments of the C-terminal extension (right).
(A) In the autoinhibited CTD RSK2 (silver), the conserved Glu500RSK2 residue from the αD-helix, which forms an ionic pair with Lys700 from the αL-helix, cannot turn toward the ATP molecule. In the active CTD MSK1 (blue), the absence of a counterpart lysine residue on the αL-helix allows repositioning of the Glu505MSK1 residue located on the αD-helix. The glutamate side chain forms a hydrogen bond (dotted line) with the ribose ring. (B) In the inactive MK2 (orange), the αL-helix has a Lys353MK2 residue, which forms an ionic pair with Glu145MK2. Similarly, in the MK2/p38 inactive complex structures (PBD codes 2OZA, 2OKR), Lys353MK2 attracts a Glu145MK2 residue from the αD-helix. (C) In contrast, the Lys353 residue of the active MK2 (yellow) is turned outward from Glu145MK2 and does not form an ionic pair. The AMP-PNP molecule is depicted from the CTD MSK1 structure and is shown in sticks (pink in A, B, C).
The C-terminal αL-helix (residues 348-363) of the inactive MK2 forms extensive hydrogen bonding contacts with the substrate-binding loop and plays an autoinhibitory role by mimicking substrate binding 26. Notably, the C-terminal αL-helix of MK2 possesses the Lys353 residue, which forms an ionic pair with Glu145MK2 from the αD-helix (Fig. 4b) similar to what was observed in the inactive RSK2. The active C-terminal truncated MK2 (residues 45-371) has the “autoinhibitory” αL-helix located at the same surface groove, but the helix lacks the network of interactions with the substrate binding loop and the αF-helix that is present in the inactive MK2. Due to an outward rotation of the αL-helix, the Lys353 of active MK2 is turned ∼90° compared to that of the inactive protein, and does not form an ionic pair with Glu145, moving away by 4 Å (Fig. 4c). The Glu505MSK1 residue conformation is similar to the Glu145MK2 in the active MK2 and differs from the corresponding glutamic acid in both the inactive MK2 and RSK2.
Potential mechanism(s) for autoinhibition of the CTD MSK1
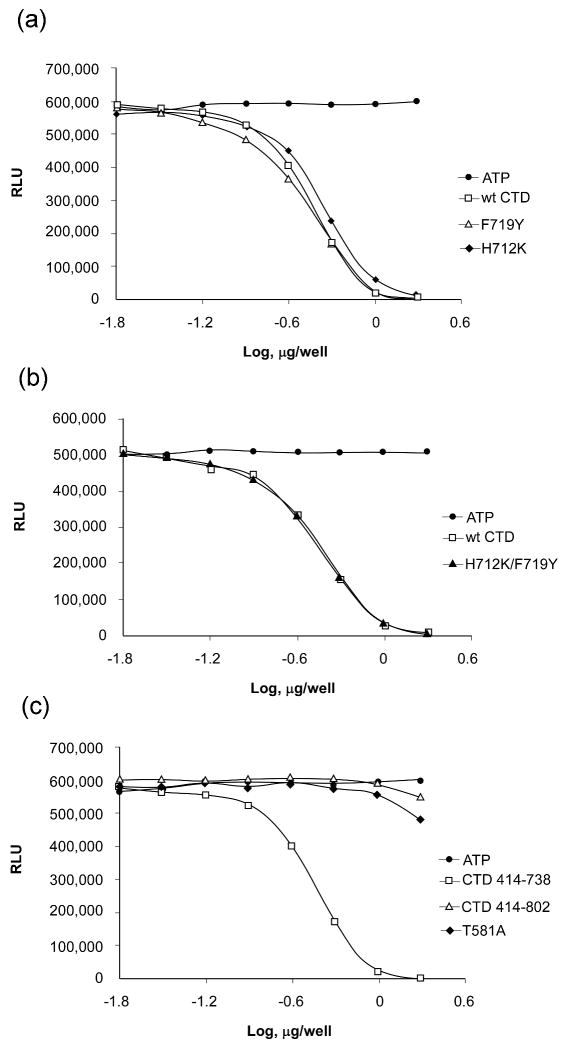
We attempted to convert the αL-helix of the CTD MSK1 to an autoinhibitory RSK2-like fragment by mutation of key residues located in the helix. His712 was mutated to lysine to create a Glu505-Lys712 salt bridge, and Phe719 was mutated to tyrosine to mimic Tyr707RSK2. However, the single point mutants, H712K and F719Y, still exhibited autophosphorylation activity comparable with the wild-type CTD MSK1 (Fig. 5a). Double mutation also had no effect on autokinase activity (Fig. 5b). The fact that the αL-helix of the CTD MSK1 cannot be converted to an autoinhibitory RSK2-like helix emphasizes a substantial dissimilarity in the role of the αL-helix in the C-terminal kinase domains of MSK1 and RSK2.
Figure 5. The kinase autophosphorylation assay for wildtype and CTD MSK1 mutants.
(A) The single point mutants, H712K and F712Y, exhibited autophosphorylation activity comparable to wildtype CTD MSK1. (B) The double mutant, H712K/F719Y, also had activity that was similar to the wildtype protein. (C) The CTD MSK1 (residues 414-738), which lacks the MAP kinase-docking site, had substantially higher activity compared with either the mutant T581A or the “full length” CTD (residues 414-802) protein. The longer CTD fragment and the T581A mutant had low residual activity and neither protein could complete the kinase reaction even up to 10 μg protein. Each experiment was repeated at least 3 times and data are expressed as Relative Luminescent Units (RLU).
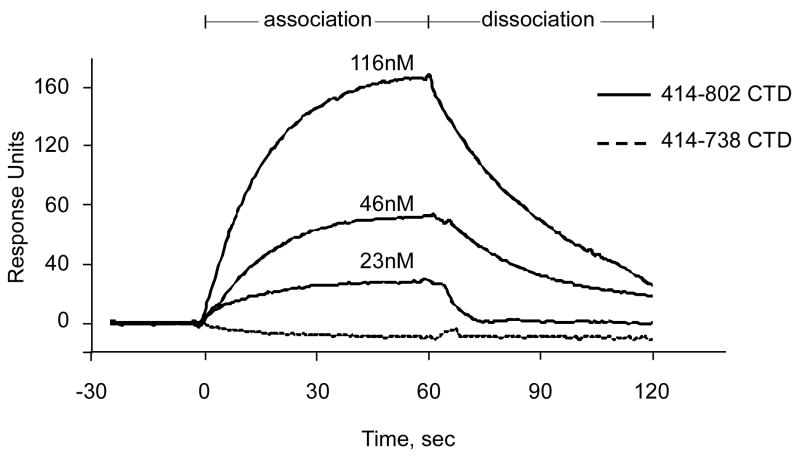
Western blot analysis indicated that the CTD MSK1 is phosphorylated at Thr581 in the T-activation loop (Supplementary Fig. S2), which could contribute to protein activity. Indeed, a purified T581A mutant showed substantially impaired autokinase activity compared with the wildtype CTD MSK1 (Fig. 5c). The disordered T-activation loop observed in several crystal structures of CTD fragments and the T581A mutant structure did not provide a definitive explanation as to how phosphorylation of Thr581 might regulate protein activity. We were interested in determining whether the deleted C-terminal end (residues 739-802), which encloses the ERK/p38 docking site, might affect the activity of the isolated CTD MSK1. The “full length” C-terminal domain of MSK1 (residues 414-802) possessed weaker autophosphorylation activity (Fig. 5c) compared with a truncated fragment (residues 414-738), which lacks the MAP kinase docking site. We performed surface plasmon resonance (SPR) binding experiments and showed that the “full length” CTD MSK1 bound with ERK1 in a dose-dependent manner with a KD of ∼ 70 nM (Fig. 6). However, the C-terminal end (residues 739-802)-deleted construct did not interact at all with ERK1 in vitro indicating that the αL-helix is not involved in the interaction with ERK.
Figure 6. Surface plasmon resonance sensorgrams.
His-tagged CTD MSK1 protein fragments were immobilized on an NTA sensor chip (GE Healthcare) as described in Experimental Procedures. The analyte, untagged ERK1 protein, was passed through the experimental and reference cells at 30 μl/min at incrementally increased concentrations (23, 46, and 116 nM). The “full length” CTD MSK1 (residues 414-802) association with ERK1, shown as increasing response units, and dissociation, shown as decreasing response units, were monitored for 60 sec. The calculated KD was 74 ± 6 nM (BIAevalation software, GE Healthcare). The CTD MSK1 containing a deleted MAP kinase-docking site (residues 414-738) did not interact with ERK1 at any concentration.
The extreme C-terminal end inhibited CTD MSK1 ex vivo kinase activity
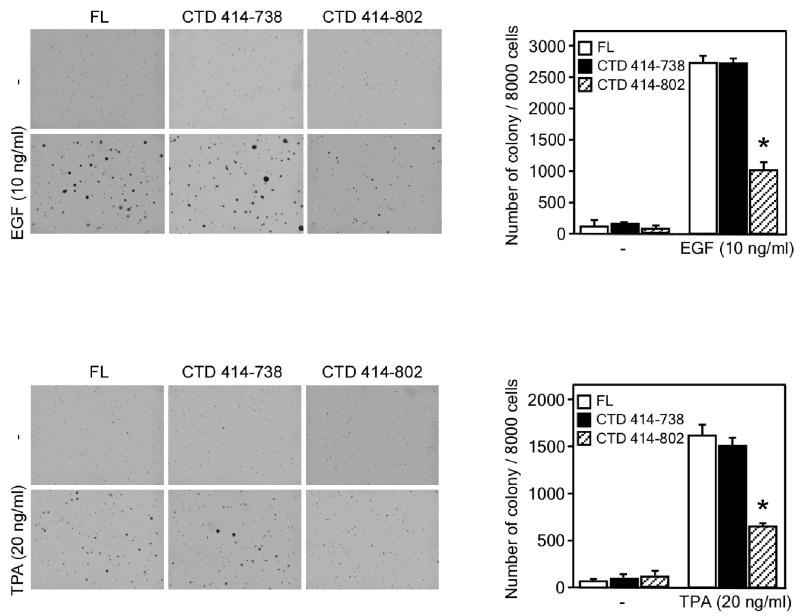
To determine whether the single C-terminal kinase domain of MSK1 has catalytic activity ex vivo, we performed a tumor promoter-induced cell transformation assay. Results indicated that overexpression of CTD MSK1 (residues 414-738) in JB6 cells resulted in neoplastic transformation in response to epidermal growth factor (EGF) or 12-O-tetradecanoyl-phorbol-13-acetate (TPA) stimulation in a manner similar to that induced by the full length MSK1 protein (Fig. 7). On the other hand, the “full length” CTD MSK1 (residues 414-802) suppressed colony formation as was previously observed for the N-terminal kinase-dead MSK1 mutant 16. This result indicates that in addition to functioning as the MAP kinase docking site, the extreme C-terminal tail (residues 739-802) is involved in the inhibition of CTD MSK1 kinase activity. The ex vivo CTD MSK1 activity was consistent with its in vitro autokinase activity.
Figure 7. Overexpression of active CTD MSK1 induces neoplastic cell transformation when stimulated with EGF or TPA.
Results of an anchorage-independent soft agar assay showed that CTD MSK1 (residues 414-738) increased EGF- (A) or TPA (B)-induced neoplastic transformation of JB6 cells in a manner similar to the wildtype MSK1 protein (FL). In contrast, the “full length” CTD MSK1 (residues 414-802) exhibited decreased cell transformation activity. The agar plates were photographed (left panels) and the average colony numbers were calculated (right panels) from 3 separate plates. Data are expressed as means ± S.D. and the asterisk indicates a significant decrease in colony formation (p < 0.05).
Discussion
Our results show that even though the CTD MSK1 has its C-terminal αL-helix in the surface groove in the same position as the corresponding helix in the autoinhibited CTD RSK2 and MK2, the CTD MSK1 displayed autophophorylation activity and was able to bind AMP-PNP in the crystals. Determination of the crystal structure of the CTD MSK1 provides us with the opportunity to observe significant differences in the interactions of the C-terminal αL-helix with the kinase domain compared to other kinases with similar structures. The autoinhibitory αL-helix of the CTD RSK2 forms hydrogen bond with the kinase core resulting in a misalignment of the conserved glutamic acid residue from the αD-helix that participates in ATP binding 20. A corresponding αL-helix in the inactive MAP kinase-activated protein kinase 2 (MK2) forms extensive hydrogen bonding with the kinase domain and mimics substrate binding 26. The crystal structure of the CTD MSK1 indicates that the C-terminal αL-helix does not form hydrogen bonds with the substrate-binding loop or nearby helices. Notably, the C-terminal αL-helix of MSK1 lacks the putative “inhibitory” residues responsible for the interaction with the kinase core. Furthermore, site-directed mutations on the αL-helix could not convert the CTD MSK1 to an autoinhibited state (i.e., CTD RSK2-like type) confirming that the αL-helix is inherently non-autoinhibitory. In contrast to what has been reported 27, the truncated MK2 does not become active because of a deletion of its “autoinhibitory” helix. The crystal structure of the active MK2 29 revealed that its C-terminal αL-helix still occupies the same “cradle” position in the surface groove; however, it lacks the hydrogen bond contacts observed in the inactive state of MK2. Our results suggest that rather than the position of the C-terminal helix itself, the network of interactions between the C-terminal extension and the catalytic kinase domain is the major determinant of autoinhibition. Previously, we suggested that the mechanism of activation for the autoinhibited CTD RSK2 involves a complex of structural re-arrangements including a displacement of the αL-helix in the surface groove, and a shift of the αD-helix with consequent re-alignment of Glu500 residue 20. Notably, the crystallized active CTD MSK1 exhibits the position of the corresponding Glu505 residue toward ATP ribose ring and its αD-helix is shifted as was predicted for the activation of CTD RSK2.
The surface plasmon resonance data directly confirmed an in vitro interaction of CTD MSK1 with the activator kinase, ERK1. The deletion of the extreme C-terminal tail (739-802 residues) encompassing the MAP kinase docking site and nuclear localization signal completely prevented the interaction with the upstream kinase, ERK1, indicating that the αL-helix is not involved in the interaction with ERK. The CTD MSK1 and MAP kinase complex might be different than that observed in the MK2/p38 complex structure 32,33. In that case, the corresponding αL-helix is located at the interface of the two proteins and contributes more than half of the buried surface area.
Cell transformation assay results indicate that the extreme C-terminal end (residues 739-802) encompassing the MAP kinase docking site might contribute to CTD MSK1 activity. We suggest that the complex formation of the CTD MSK1 with phosphorylating kinases occurs through the MAP kinase docking site without the involvement of the αL-helix, and triggers conformational changes analogous to an artificial deletion of the C-terminal tail resulting in protein activation. Overall we have shown that the C-terminal αL-helix of CTD MSK1 does not play a dominant role in autoinhibition in contrast to CTD RSK2, which is activated in an αL-helix-mediated manner. The CTD MSK1 structure suggests that diverse mechanisms of autoinhibition and activation might be adopted by members of this closely related protein kinase family.
Materials and Methods
Expression and purification of soluble CTD MSK1 fragments
Eleven different constructs of CTD MSK1 comprising residues 398-802 were subcloned into the NdeI/XhoI restriction sites of the pET-28a vector (Novagen) using full length human MSK1 (GenBank AF074393.1) as a template. The recombinant CTDs MSK1 were expressed in E. coli BL21- Codon Plus (DE3)-RIPL competent cells (Stratagene). The cells were induced with 0.25 mM isopropyl-β-D-thiogalactopyranoside and then harvested after an additional 5 h of growth at 25 °C. The cells were disrupted by French press (Thermo Electron Corp.) in buffer (30 mM imidazole, 500 mM NaCl, 50 mM sodium phosphate, pH 8.0, 10% glycerol, 10 mM β-mercaptoethanol). The soluble His-tagged CTD MSK1 proteins were purified on nickel-nitrilotriacetic acid agarose (Ni-NTA; Qiagen) and eluted with an increasing imidazole step gradient (100 and 250 mM). All fractions were loaded onto a HiLoad 16/60 Superdex-200 column (GE Healthcare). For most of the CTD constructs, size exclusion chromatography showed a peak that corresponded to a dimer by elution time and was confirmed by SDS-PAGE as a double band. Three constructs comprising residues 398-738, 414-738, and 414-750 eluted from the gel-filtration column as a single peak with a shoulder. The peak position corresponded to a dimer and showed a double band on SDS-PAGE. The shoulder fractions corresponded to a monomer by elution time. The monomeric fractions of the shoulder confirmed by SDS-PAGE as a single band were collected, concentrated and re-loaded onto a Superdex-200 gel-filtration column and used for crystallization. Site-directed mutagenesis was performed using a QuickChange® Lightning site-directed mutagenesis kit (Stratagene). Mutated proteins were expressed and purified by a similar procedure and exhibited an equal yield and purity to wildtype. Full length human ERK1 (Swiss-Prot P27361.4) was cloned into the pET-46 Ek/LIC vector (Novagen). The His-tagged ERK1 was purified on Ni-NTA and treated with enterokinase to remove the His-tag followed by size-exclusion chromatography on Superdex-200. Purification of the CTD RSK2 (399-740 residues) was performed as described previously 20.
Crystallization and data collection
The monomeric fractions of the only three constructs mentioned above were found to produce crystals. “Full length” CTD MSK1 (residues 414-802) did not produce crystals. The best diffraction data were obtained for the CTD MSK1 construct encompassing residues 414-738. The protein was concentrated to 10-20 mg/ml in buffer composed of 150 mM NaCl, 20 mM Tris (pH 8.0), and 10 mM β-mercaptoethanol. The diluted protein (2.5-5 mg/ml) was mixed with precipitant at a 1:1 ratio. The precipitant solution was composed of 20% PEG 3350, 0.1-0.2 M ammonium acetate, and 0.1 M Hepes (pH 7.5). The crystals were grown at 20 °C by the sitting drop vapor diffusion method. Single and clusters of thin needle and thin plate-like crystals appeared in 1 wk and were grown over the next 2-3 wks. The crystals were transferred to a cryo-protectant solution (20% PEG 3350, 0.1-0.2 M ammonium acetate, 0.1 M Hepes pH 7.5, 20% ethylene glycol), and flash-cooled in liquid nitrogen. The full diffraction data sets were collected from several portions of the thin plate-like crystals at the 24ID-E, 24ID-C beamlines of the Advanced Photon Source (APS). The best data for the AMP-PNP complex were obtained when the native crystals were soaked with 5 mM AMP-PNP and 5 mM MgCl2 for 5 hours.
Structure determination
Data were processed using HKL2000 34 and molecular replacement was performed using PHASER 35. The structure of the ligand-free C-terminal kinase domain of p90 ribosomal S6 kinase 2 (RSK2) (PDB code 2QR8) with all waters deleted was used as a search model for CTD MSK1. The highest scoring solution placed a homodimer in the asymmetric unit that was used as a starting model for re-building and structure refinement, which was initially performed using PHENIX 36 (initial autorebuild), CNS 37 (torsional and cartesian simulated annealing), and REFMAC (CCP4 suite, 38 in alternation with manual rebuilding using COOT 39. During refinement, tight NCS restraints were maintained on the homodimer. For the CTD MSK1/AMP-PNP complex, the apo form of CTD MSK1 was used to calculate phases. Difference Fourier electron density maps revealed residual density within the active site consistent with a bound AMP-PNP molecule. A model for the substrate was established using the PRODRG server 40. Electron density maps were calculated with the CCP4 suite (FFT function). The final models of the apo form and the CTD MSK1/AMP-PNP complex contained a homodimer encompassing residues 415-729. Electron density corresponding to one molecule of AMP-PNP was readily identified in each protein molecule. The magnesium cation was not visible in the electron density map. Residues 555-558 and the T-activation loop (residues 575-596), including the primary phosphorylation site Thr581 were disordered and not visible in either the ligand-bound or apo form of CTD MSK1. A short segment, spanning residues 624-630, was only visible in chain B (modeled as polyalanine in the apo CTD MSK1). The two molecules in the asymmetric unit were very similar in both forms of CTD MSK1 (r.m.s.d. 0.44 Å for the 279 corresponding Cα atoms), with a slight movement of the C-lobe and a re-arrangement of the loop comprising residues 669-675. We observed significantly higher B factors in the CTD MSK1/AMP-PNP complex compared to the apo form (53.4 Å2 vs 27.8 Å2). This was most likely due to more disordered areas and a somewhat lower crystal quality. In addition, the complex protein, albeit being very similar to its ligand-free counterpart, contained more flexible areas, particularly in the loop regions. The dimer interface and buried surface calculations were performed using the program SPPIDER 41.
The CTD MSK1 crystal structures of the longer construct (residues 414-750, resolution 2.4 Å) and the T581A mutant (resolution 2.2 Å) were determined using molecular replacement. Both structures were highly isomorphous to the wildtype, with a maximum variation in unit cell axis of less than 1%, but did not provide additional insight due to disordered regions at the C-terminus (construct 414-750) and around the Thr581 residue (within the T activation loop). Structural figures and graphical rendering were made using PYMOL (http://pymol.sourceforge.net).
Luminescent autophosphorylation assay
The Kinase-Glo® Luminescent Assay (Promega Corp.) was performed in a single well (reaction volume of 50 μl) of a 96-well plate according to the manufacturer's instructions. Twofold serial dilutions of the CTD fragments were made across the plate in kinase reaction buffer (40 mM Tris, pH 7.5, 20 mM MgCl2, 0.1 mg/ml BSA, 0.1 μM ATP). The kinase reaction was run for 45 min at room temperature and terminated by the addition of 50 μl of Kinase-Glo® Reagent. The luminescence signal was recorded on the Luminoscan plate reader (Labsystem) 10 min later. All experiments were reproduced several times.
Surface plasmon resonance experiments (SPR)
The ligand (His-tagged CTD MSK1; 15 ng) was immobilized on an NTA sensor chip and equilibrated in HBS-P buffer that contained 150 mM NaCl, 10 mM Hepes pH 7.5, 50 μM EDTA, and 0.01% P-20. An analyte (non-tagged ERK1) was passed through experimental and reference cells at 30 μl/min at incrementally increased concentrations. The association was monitored for 60 sec as an increase of response units. The experiments were performed using the Biacore® X (GE Healthcare). The KD value was calculated using BIAevalation software (GE Healthcare).
Stable cell lines and cell transformation assay
Full length MSK1 and two different CTD constructs (residues 414-738 and 414-802) were cloned into the pcDNA4A vector and transfected into JB6 CI41 mouse epidermal skin cells. Stable cell lines were selected in medium containing 600 μg/mL zeocin as described previously 16. Cells (8 × 103) were subjected to a soft agar assay in the presence of EGF (10 ng/ml) or TPA (20 ng/ml) in 1 ml of 0.3% BME agar containing 10% FBS. The cultures were maintained at 37 °C in a 5% CO2 atmosphere for 10-12 days and then colonies were counted using a microscope and the Image-Pro PLUS (v.4) computer software program. The expression level of MSK1 proteins was confirmed by Western blot analysis.
Western blot analysis
Samples of purified CTD MSK1 containing 0.2 or 0.4 μg of protein were resolved by SDS-PAGE and transferred onto PVDF membranes. The membranes were incubated in blocking buffer and hybridized with a specific antibody to detect phosphorylated Thr581 (Cell Signaling). The Western blots were visualized using an enhanced chemiluminescence detection system after hybridization with a horseradish peroxidase-conjugated secondary antibody.
Accession codes
Structure factors and coordinates of CTD MSK1 in apo form and in complex with AMP-PNP have been deposited in the Protein Data Bank with PBD ID: 3KN6 and 3KN5, respectively.
Supplementary Material
The dimerization of theCTD MSK1 occurs through a network of hydrogen contacts between the αD/αK-helix junction from one CTD molecule and the loop preceding the αK-helix from another CTD protein. The dimerization interface, mainly composed of charged residues, Lys512, Lys514, Asn629, Asn655, and Glu508, buries an area of ∼ 300 Å2. The αK- and αL-helices are shown in red.
The purified CTD MSK1 protein was subjected to Western blotting using a specific antibody to detect phosphorylation of the Thr581 residue.
Acknowledgments
The work was funded by The Hormel Foundation and NIH grants CA027502, CA077646, CA120388, R37CA08164 and ES016548. A part of the work is based upon research conducted at the Northeastern Collaborative Access Team beamlines 24ID of the Advanced Photon Source (APS) and was supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the APS is supported by the US Department of Energy, Office of Basic Energy Sciences, under contract No. DE-AC02-06CH11357. Part of the work was conducted at the Canadian Light Source (CLS-CMCF).
Abbreviations used
- CTD MSK1
the C-terminal kinase domain of the mitogen- and stress-activated protein kinase 1
- RSK2
p90 ribosomal S6 protein kinase 2
- MAP kinase
mitogen-activated protein kinase
- PDB
protein data bank
- PEG
polyethylene glycol
- Ni-NTA
nickel-nitrilotriacetic acid agarose
Footnotes
Competing interests statement
Authors declare that they have no conflict of interest.
Supplementary data include two figures and can be found in the online version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Deak M, Clifton AD, Lucocq LM, Alessi DR. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. Embo J. 1998;17:4426–41. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierrat B, Correia JS, Mary JL, Tomas-Zuber M, Lesslauer W. RSK-B, a novel ribosomal S6 kinase family member, is a CREB kinase under dominant control of p38alpha mitogen-activated protein kinase (p38alphaMAPK) J Biol Chem. 1998;273:29661–71. doi: 10.1074/jbc.273.45.29661. [DOI] [PubMed] [Google Scholar]
- 3.Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell. 2006;24:367–81. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–15. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 5.Duncan EA, Anest V, Cogswell P, Baldwin AS. The kinases MSK1 and MSK2 are required for epidermal growth factor-induced, but not tumor necrosis factor-induced, histone H3 Ser10 phosphorylation. J Biol Chem. 2006;281:12521–5. doi: 10.1074/jbc.M513333200. [DOI] [PubMed] [Google Scholar]
- 6.Arthur JS, Cohen P. MSK1 is required for CREB phosphorylation in response to mitogens in mouse embryonic stem cells. FEBS Lett. 2000;482:44–8. doi: 10.1016/s0014-5793(00)02031-7. [DOI] [PubMed] [Google Scholar]
- 7.Wiggin GR, Soloaga A, Foster JM, Murray-Tait V, Cohen P, Arthur JS. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol Cell Biol. 2002;22:2871–81. doi: 10.1128/MCB.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck IM, Vanden Berghe W, Vermeulen L, Bougarne N, Vander Cruyssen B, Haegeman G, De Bosscher K. Altered subcellular distribution of MSK1 induced by glucocorticoids contributes to NF-kappaB inhibition. Embo J. 2008;27:1682–93. doi: 10.1038/emboj.2008.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janknecht R. Regulation of the ER81 transcription factor and its coactivators by mitogen- and stress-activated protein kinase 1 (MSK1) Oncogene. 2003;22:746–55. doi: 10.1038/sj.onc.1206185. [DOI] [PubMed] [Google Scholar]
- 10.Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. Embo J. 2003;22:2788–97. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthur JS, Fong AL, Dwyer JM, Davare M, Reese E, Obrietan K, Impey S. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J Neurosci. 2004;24:4324–32. doi: 10.1523/JNEUROSCI.5227-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darragh J, Soloaga A, Beardmore VA, Wingate AD, Wiggin GR, Peggie M, Arthur JS. MSKs are required for the transcription of the nuclear orphan receptors Nur77, Nurr1 and Nor1 downstream of MAPK signalling. Biochem J. 2005;390:749–59. doi: 10.1042/BJ20050196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuck S, Soloaga A, Schratt G, Arthur JS, Nordheim A. The kinase MSK1 is required for induction of c-fos by lysophosphatidic acid in mouse embryonic stem cells. BMC Mol Biol. 2003;4:6. doi: 10.1186/1471-2199-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ananieva O, Darragh J, Johansen C, Carr JM, McIlrath J, Park JM, Wingate A, Monk CE, Toth R, Santos SG, Iversen L, Arthur JS. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat Immunol. 2008;9:1028–36. doi: 10.1038/ni.1644. [DOI] [PubMed] [Google Scholar]
- 15.Cohen P. Targeting protein kinases for the development of anti-inflammatory drugs. Curr Opin Cell Biol. 2009;21:317–24. doi: 10.1016/j.ceb.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Kim HG, Lee KW, Cho YY, Kang NJ, Oh SM, Bode AM, Dong Z. Mitogen- and stress-activated kinase 1-mediated histone H3 phosphorylation is crucial for cell transformation. Cancer Res. 2008;68:2538–47. doi: 10.1158/0008-5472.CAN-07-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCoy CE, Campbell DG, Deak M, Bloomberg GB, Arthur JS. MSK1 activity is controlled by multiple phosphorylation sites. Biochem J. 2005;387:507–17. doi: 10.1042/BJ20041501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy CE, macdonald A, Morrice NA, Campbell DG, Deak M, Toth R, McIlrath J, Arthur JS. Identification of novel phosphorylation sites in MSK1 by precursor ion scanning MS. Biochem J. 2007;402:491–501. doi: 10.1042/BJ20061183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KJ, Carter PS, Bridges A, Horrocks P, Lewis C, Pettman G, Clarke A, Brown M, Hughes J, Wilkinson M, Bax B, Reith A. The structure of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein. Structure. 2004;12:1067–77. doi: 10.1016/j.str.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 20.Malakhova M, Tereshko V, Lee SY, Yao K, Cho YY, Bode A, Dong Z. Structural basis for activation of the autoinhibitory C-terminal kinase domain of p90 RSK2. Nat Struct Mol Biol. 2008;15:112–3. doi: 10.1038/nsmb1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-Levy R, Hooper S, Wilson R, Paterson HF, Marshall CJ. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr Biol. 1998;8:1049–57. doi: 10.1016/s0960-9822(98)70442-7. [DOI] [PubMed] [Google Scholar]
- 22.Engel K, Kotlyarov A, Gaestel M. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. Embo J. 1998;17:3363–71. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomas-Zuber M, Mary JL, Lamour F, Bur D, Lesslauer W. C-terminal elements control location, activation threshold, and p38 docking of ribosomal S6 kinase B (RSKB) J Biol Chem. 2001;276:5892–5899. doi: 10.1074/jbc.M005822200. [DOI] [PubMed] [Google Scholar]
- 24.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–44. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 26.Meng W, Swenson LL, Fitzgibbon MJ, Hayakawa K, Ter Haar E, Behrens AE, Fulghum JR, Lippke JA. Structure of mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 suggests a bifunctional switch that couples kinase activation with nuclear export. J Biol Chem. 2002;277:37401–5. doi: 10.1074/jbc.C200418200. [DOI] [PubMed] [Google Scholar]
- 27.Underwood KW, Parris KD, Federico E, Mosyak L, Czerwinski RM, Shane T, Taylor M, Svenson K, Liu Y, Hsiao CL, Wolfrom S, Maguire M, Malakian K, Telliez JB, Lin LL, Kriz RW, Seehra J, Somers WS, Stahl ML. Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme. Structure. 2003;11:627–36. doi: 10.1016/s0969-2126(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 28.Hillig RC, Eberspaecher U, Monteclaro F, Huber M, Nguyen D, Mengel A, Muller-Tiemann B, Egner U. Structural basis for a high affinity inhibitor bound to protein kinase MK2. J Mol Biol. 2007;369:735–45. doi: 10.1016/j.jmb.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Anderson DR, Meyers MJ, Vernier WF, Mahoney MW, Kurumbail RG, Caspers N, Poda GI, Schindler JF, Reitz DB, Mourey RJ. Pyrrolopyridine inhibitors of mitogen-activated protein kinase-activated protein kinase 2 (MK-2) J Med Chem. 2007;50:2647–54. doi: 10.1021/jm0611004. [DOI] [PubMed] [Google Scholar]
- 30.Poteet-Smith CE, Smith JA, Lannigan DA, Freed TA, Sturgill TW. Generation of constitutively active p90 ribosomal S6 kinase in vivo. Implications for the mitogen-activated protein kinase-activated protein kinase family. J Biol Chem. 1999;274:22135–8. doi: 10.1074/jbc.274.32.22135. [DOI] [PubMed] [Google Scholar]
- 31.Kang S, Elf S, Dong S, Hitosugi T, Lythgoe K, Guo A, Ruan H, Lonial S, Khoury HJ, Williams IR, Lee BH, Roesel JL, Karsenty G, Hanauer A, Taunton J, Boggon TJ, Gu TL, Chen J. Fibroblast growth factor receptor 3 associates with and tyrosine phosphorylates p90 RSK2, leading to RSK2 activation that mediates hematopoietic transformation. Mol Cell Biol. 2009;29:2105–17. doi: 10.1128/MCB.00998-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ter Haar E, Prabhakar P, Liu X, Lepre C. Crystal structure of the p38 alpha-MAPKAP kinase 2 heterodimer. J Biol Chem. 2007;282:9733–9. doi: 10.1074/jbc.M611165200. [DOI] [PubMed] [Google Scholar]
- 33.White A, Pargellis CA, Studts JM, Werneburg BG, Farmer BT., 2nd Molecular basis of MAPK-activated protein kinase 2:p38 assembly. Proc Natl Acad Sci U S A. 2007;104:6353–8. doi: 10.1073/pnas.0701679104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–82. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project, N.T. C. S. P. f. P. C. Acta Crystallographica. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des. 1996;10:255–62. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 41.Porollo A, Meller J. Prediction-based fingerprints of protein-protein interactions. Proteins. 2007;66:630–45. doi: 10.1002/prot.21248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The dimerization of theCTD MSK1 occurs through a network of hydrogen contacts between the αD/αK-helix junction from one CTD molecule and the loop preceding the αK-helix from another CTD protein. The dimerization interface, mainly composed of charged residues, Lys512, Lys514, Asn629, Asn655, and Glu508, buries an area of ∼ 300 Å2. The αK- and αL-helices are shown in red.
The purified CTD MSK1 protein was subjected to Western blotting using a specific antibody to detect phosphorylation of the Thr581 residue.