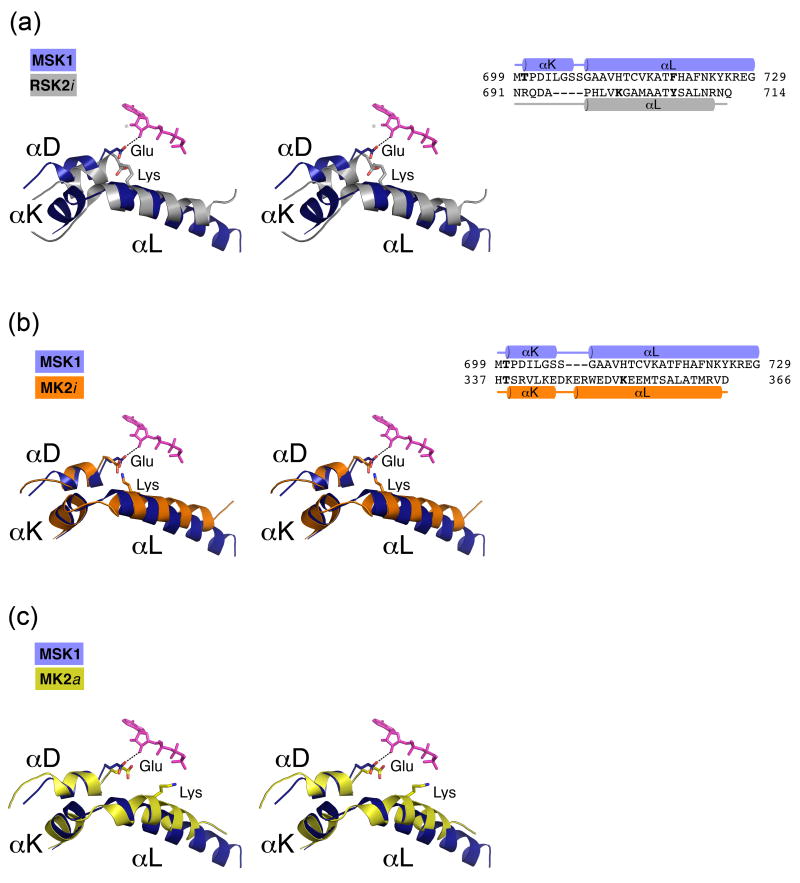
Figure 4. An enlarged view of the αL/αD helix interaction (left) and the 3D-structure based alignments of the C-terminal extension (right).
(A) In the autoinhibited CTD RSK2 (silver), the conserved Glu500RSK2 residue from the αD-helix, which forms an ionic pair with Lys700 from the αL-helix, cannot turn toward the ATP molecule. In the active CTD MSK1 (blue), the absence of a counterpart lysine residue on the αL-helix allows repositioning of the Glu505MSK1 residue located on the αD-helix. The glutamate side chain forms a hydrogen bond (dotted line) with the ribose ring. (B) In the inactive MK2 (orange), the αL-helix has a Lys353MK2 residue, which forms an ionic pair with Glu145MK2. Similarly, in the MK2/p38 inactive complex structures (PBD codes 2OZA, 2OKR), Lys353MK2 attracts a Glu145MK2 residue from the αD-helix. (C) In contrast, the Lys353 residue of the active MK2 (yellow) is turned outward from Glu145MK2 and does not form an ionic pair. The AMP-PNP molecule is depicted from the CTD MSK1 structure and is shown in sticks (pink in A, B, C).