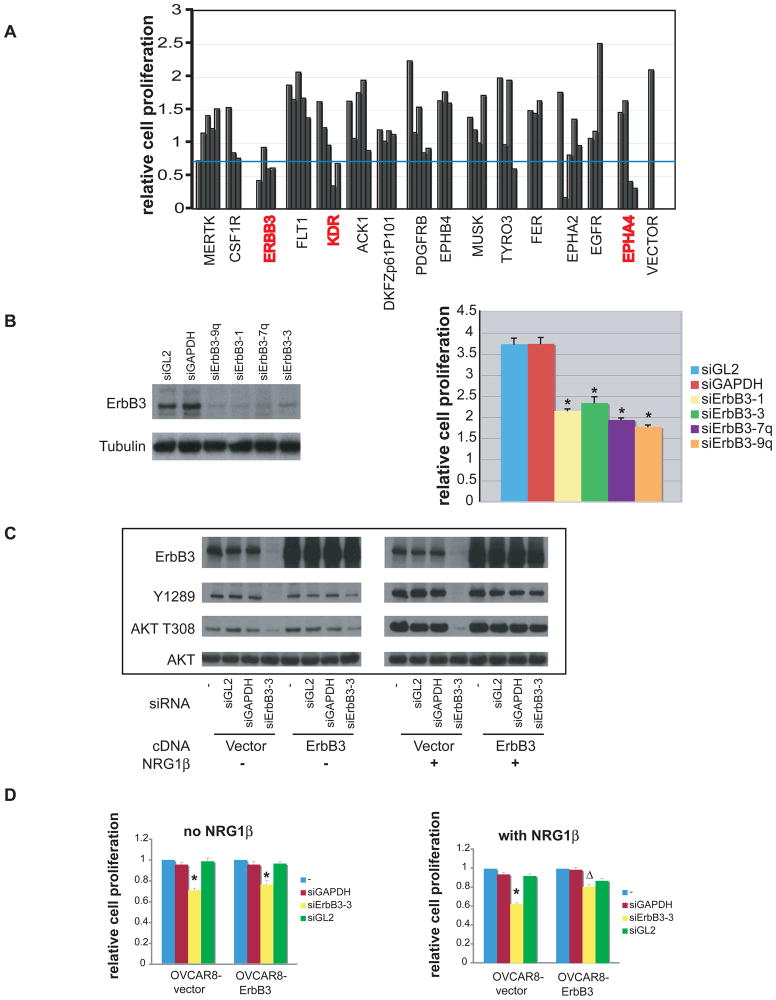
Figure 1. Depletion of ErbB3 inhibits OVCAR8 proliferation.
A) Secondary shRNA kinase screen in which 15 hairpin sets, each of which scored at least one hit in the primary screen, were retested. Each bar represents the effect of one hairpin; shRNAs directed against the same kinase are grouped together. Hairpin sets containing ≥2 hits in the secondary screen were considered positives in that screen and marked in red. The blue line denotes the point representing one standard deviation below the mean value of all CellTiter-Glo data obtained in this assay. See Table S1 for complete results of the screen. B) SiRNA species directed against ErbB3 inhibited OVCAR8 proliferation. Left panel, ErbB3 protein level in OVCAR8 cells treated with either control siRNA or different siErbB3 species. Right panel, proliferation of OVCAR8 treated with either control siRNA or siErbB3. *: significantly different from siGL2 or siGAPDH treated sample (p<0.05, student’s t–test. See also Fig. S1 for effects of shErbB3. C) Biochemical effects of over-expressing an ErbB3 cDNA immune to ErbB3 siRNA in the absence and presence of recombinant NRG1β (see Materials and Methods). D) Effect on OVCAR8 proliferation of over-expressing an ErbB3 cDNA immune to ErbB3 siRNA in the absence and presence of NRG1β. In the vector-only control cells, growth of ErbB3 siRNA cells was significantly different from siGL2 and siGAPDH treated samples (*, p<0.05, student’s t-test). In cells stably expressing siRNA-resistant ErbB3 cDNA in the presence of exogenous NRG-1β, growth of cells treated with ErbB3 siRNA was significantly greater than in the vector only cells (Δ, p<0.05, student’s t-test). Error bars represent +/− SD.