Abstract
The methylotrophic yeast Pichia pastoris is widely used for the expression of heterologous enzymes. While the purity of the desired expression product is of major importance for many applications, we found that recombinant enzymes produced in methanol medium were contaminated by a 37-kDa endogenous yeast protease. This enzyme was completely inhibited by phenylmethanesulfonyl fluoride (PMSF) but not by 1,10-phenanthroline, EDTA, and pepstatin A, suggesting the nature of a serine protease. Its secretion was abolished in P. pastoris strains GS115 and KM71 by specific mutagenesis of a subtilisin gene (SUB2) but not by inactivation of the gene encoding vacuolar proteinase B (PRB). Bioinformatic comparisons of Sub2 protein with subtilisins from other fungal genomes and phylogenetic analyses indicated that this enzyme is not an orthologue of the vacuolar protease cerevisin generally present in yeasts but is more closely related to another putative subtilisin found in a small number of yeast genomes. During growth of P. pastoris, Sub2 was produced as a secreted enzyme at a concentration of 10 μg/ml of culture supernatant after overexpression of the full-length SUB2 gene. During fermentative production of recombinant enzymes in methanol medium, 1 ml of P. pastoris culture supernatant was found to contain approximately 3 ng of Sub2, while the enzyme was not detected during growth in a medium containing glycerol as a carbon source. The mutant strain GS115-sub2 was subsequently used as a host for the production of recombinant proteases without endogenous subtilisin contamination.
The methylotrophic yeast Pichia pastoris has been used successfully to express a wide range of heterologous proteins. Pichia pastoris has two genes that encode alcohol oxidase, AOX1 and AOX2. The transcription of the former gene is tightly regulated by methanol, while the latter is expressed in small quantities (7, 11). Alcohol oxidase is not found in the cell when P. pastoris grows in the presence of glycerol, glucose, or ethanol, but in the presence of methanol, alcohol oxidase enzyme 1 (Aox1) amounts to 5% of total cellular proteins in shake-flask cultures and over 30% of total cellular proteins in fermenter cultures (6). The procedure for producing a secreted recombinant protein by using P. pastoris consists of cloning the cDNA encoding the protein of interest downstream of a signal sequence under the control of the AOX1 promoter in a P. pastoris expression vector. In general, the P. pastoris acid phosphatase gene (PHO1) signal sequence or the α-factor signal peptide sequence is used for entry of the secretory pathway of the yeast (19). The construct, which carries in addition to the cloned coding sequence of interest a gene for selection after transformation of P. pastoris, is inserted into the P. pastoris genome at the AOX1 locus via homologous recombination. Selected transformants are screened for recombinant protein production after induction in a medium containing methanol.
A fundamental objective of our research on fungi (Aspergillus spp. and dermatophytes) is to gain a comprehensive view of the enzymes that allow the digestion of an insoluble protein structure, such as keratinized tissues, into oligopeptides and free amino acids (32, 33). Highly purified proteases are needed to analyze the different steps in protein and peptide digestions. However, purification of an individual protease from fungal culture supernatant and cell extract is a laborious process, especially when the protease of interest is produced in small amounts and simultaneously with other proteases. Therefore, P. pastoris is a powerful tool for individual production of numerous secreted proteases as recombinant proteins in substantial amounts for further characterization and applications (1, 2, 4, 24, 31, 34, 41).
During assays of sequential digestion of large peptides by exoproteases, we realized that purified fractions of recombinant leucine aminopeptidases, dipeptidyl peptidases, and tripeptidyl peptidases of the sedolisin family after gel filtration and ion-exchange chromatography had additional endoproteolytic activities that were similar in all trials. In the present study, we identified and characterized a contaminating protease of the subtilisin family which is secreted in minor amounts by P. pastoris during growth in methanol medium. A strain deficient in this endogenous secreted subtilisin activity was constructed for further heterologous gene expression experiments.
MATERIALS AND METHODS
Strains and plasmids.
Pichia pastoris GS115 and KM71 (Invitrogen, Carlsbad, CA), pPICZA, and previously constructed plasmids pKJ113-LAP2 and pKJ113-DPPIV for expression of the genes encoding Aspergillus fumigatus DppIV (AfuDppIV) and A. fumigatus Lap2 (AfuLap2), respectively, were used (2, 31). Escherichia coli LE392 was used for the propagation of the bacteriophage λEMBL3 (Promega, Madison, WI). Direct cloning of PCR products in the pDrive vector was performed using a PCR cloning kit (Qiagen, Hombrechtikon, Switzerland). All plasmid subcloning experiments were performed with E. coli XL1-Blue with plasmids pUC19 and pPICZA (Invitrogen).
Cloning of the P. pastoris gene encoding PrB.
A P. pastoris genomic λEMBL3 library was constructed as previously described for Candida parapsilosis (10). Saccharomyces cerevisiae DNA encoding vacuolar proteinase B (PrB) was used as a probe for cloning of the orthologous gene in P. pastoris. Recombinant plaques (2 × 104) of the genomic library were immobilized on GeneScreen nylon membranes (NEN Life Science Products). The filters were hybridized with 32P-labeled probe under low-stringency conditions (30). All positive plaques were purified, and the associated bacteriophage DNAs were isolated as described previously (16). Agarose gel electrophoresis of restricted recombinant bacteriophage λEMBL3 DNA, Southern blotting, and subcloning of hybridizing fragments from bacteriophages into pUC19 were performed using standard protocols (36). DNA was sequenced by Microsynth (Balgach, Switzerland).
Pichia pastoris transformation.
Pichia pastoris strains were transformed by electroporation with 5 to 10 μg of linearized plasmid DNA. Using pPICZA construct DNA, transformants were selected on YPDS medium containing 100 μg ml−1 Zeocin (Invitrogen). His+ GS115 and GS115-sub2 transformants were selected on histidine-deficient medium (1 M sorbitol, 1% [wt/vol] dextrose, 1.34% [wt/vol] yeast nitrogen base [YNB] without amino acids, 4 × 10−5% [wt/vol] biotin, 5 × 10−3% amino acids [i.e., 5 × 10−3% {wt/vol} each l-glutamic acid, l-methionine, l-lysine, l-leucine, and l-isoleucine], 2% [wt/vol] agarose). Subsequently, the transformants were screened for insertion of the construct at the AOX1 site on minimal-methanol plates (1.34% [wt/vol] YNB without amino acids, 4 × 10−5% [wt/vol] biotin, 0.5% [vol/vol] methanol, 2% [wt/vol] agarose). Transformants unable to grow on media containing only methanol as a carbon source were retained for further investigations. They were assumed to contain the construct at the correct yeast genomic location by integration events in the AOX1 locus displacing the AOX1 coding region.
Native secreted subtilisin and heterologous enzyme production in P. pastoris.
Pichia pastoris GS115 and KM71 as well as P. pastoris transformants were grown to near saturation (optical density [OD] of 20 at 600 nm) at 30°C in 10 ml of glycerol-based yeast media (0.1 M potassium phosphate buffer at pH 6.0, containing 1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 1.34% [wt/vol] YNB without amino acids, 1% [vol/vol] glycerol, and 4 × 10−5% [wt/vol] biotin). Cells were harvested by centrifugation and resuspended in 2 ml of the same medium with 0.5% (vol/vol) methanol instead of glycerol and incubated for 2 days. Thereafter, the culture supernatants were separated from the yeast cells by centrifugation and retained for protein analysis. Salts and low-molecular-weight solutes were removed from 2.5 ml of P. pastoris culture supernatant by passing through a PD10 column (Amersham Pharmacia, Buckinghamshire, United Kingdom) by using 20 mM Tris-HCl buffer (pH 7.5) before testing for proteolytic activity. Protein concentrations were measured by the Bradford method with a commercial reagent (Bio-Rad, Hercules, CA) and by densitometry on SDS-PAGE gels with the aid of Adobe Photoshop software (Adobe Systems Inc., San Jose, CA) with different amounts of bovine serum albumin as standards.
Protein gel electrophoresis.
Extracts were analyzed by SDS-PAGE (25) with a separation gel of 12% polyacrylamide. To detect proteolytic activity of culture supernatant, gelatin at a concentration of 0.1% was added to the gel. In this case, 10 μl of P. pastoris culture supernatant was simply added to an equal volume of SDS-PAGE sample buffer without β-mercaptoethanol (125 mM Tris, pH 6.8, 4% SDS, 20% glycerol, 0.002% bromophenol blue) and the mixture was loaded without heating onto SDS-PAGE gels. Samples were pretreated with phenylmethanesulfonyl fluoride (PMSF) or EDTA at a concentration of 10 or 200 mM, respectively, during 10 min at room temperature before the samples were loaded for the characterization of proteolytic activities. After electrophoresis, the gels were incubated twice for 15 min at room temperature in 10 volumes of 2.5% Triton X-100 under constant agitation. The gels were then incubated overnight (16 h) with 10 mM sodium phosphate buffer, pH 7.0 or pH 4.0, in the presence of or in the absence of PMSF or EDTA when the samples had been treated with these proteolytic inhibitors before loading. The gels were stained for 30 min with 0.1% Coomassie brilliant blue R-250 (Bio-Rad) in ethanol-acetic acid-water (40:10:50) and destained with the same solvent.
Proteolytic activity.
Proteolytic activity was assayed using resorufin-labeled casein as previously described (5) and the chromogenic substrate N-succinyl-Ala-Ala-Pro-Phe-p-nitrounilide (N-Suc-Ala-Ala-Pro-Phe-pNA), (Sigma) (8, 28). Sodium acetate buffer (pH 3 to 6.5), Tris-HCl buffer (pH 7 to 9), and glycine-NaOH buffer (pH 9 to 11) were used. With N-Suc-Ala-Ala-Pro-Phe-pNA, a substrate stock solution was prepared at a 20 mM concentration (12 mg/ml) in dimethyl sulfoxide (DMSO) and stored at −20°C. The reaction mixture contained a concentration of 1.0 mM substrate and enzyme preparation (between 0.1 to 1.0 μg per assay) in 100 μl of 50 mM buffer, at different pH values. After incubation at 37°C for 60 min, the reaction was terminated by the addition of 5 μl of glacial acetic acid followed by the addition of 0.9 ml of water to the mixture. The released pNA was measured by spectrometry as the change in A405. A control with a blank substrate and blank culture broth was carried out in parallel. The enzyme activities were expressed in mU (nmol of released pNA/min at pH 7.0) with N-Suc-Ala-Ala-Pro-Phe-pNA as a substrate.
Inhibitors were from a protease inhibitor set (Roche Diagnostics, Rotkreuz, Switzerland), and stock solutions were prepared as recommended by the supplier. Sub2 (0.1 μg) in 0.2 ml of 20 mM Tris-HCl buffer (pH 7.0) was incubated for 10 min at 37°C either with PMSF (10 μg/ml) or with antipain (50 μg/ml), bestatin (40 μg/ml), chymostatin (6 μg/ml), E64 (10 μg/ml), leupeptin (5 μg/ml), pepstatin (0.7 μg/ml), phosphoramidon (330 μg/ml), EDTA (0.2 mg/ml), or aprotinin (2 μg/ml), following the recommendations of the supplier. The mixture was incubated for 10 min at 37°C, and assays were subsequently performed after the addition of the substrate to the mixture. An appropriate control without inhibitor was assayed simultaneously.
Protein identification by liquid chromatography-tandem mass spectrometry (LC-MS-MS).
Coomassie blue-stained bands were excised from the SDS-PAGE gels and in-gel digested with sequencing-grade trypsin (Promega) as previously described (37, 40). Extracted peptides were analyzed on a hybrid linear trap LTQ Orbitrap mass spectrometer (Thermo Fisher, Bremen, Germany) interfaced via a TriVersa Nanomate (Advion Biosciences, Norwich, United Kingdom) to an Agilent 1100 nano-high-performance-liquid-chromatography (nano-HPLC) system (Agilent Technologies, Waldbronn, Germany). Solvents used for the mobile phase were 95:5 H2O-acetonitrile (vol/vol) with 0.1% formic acid (solvent A) and 5:95 H2O-acetonitrile (vol/vol) with 0.1% formic acid (solvent B).
Peptides were loaded onto a ZORBAX 300SB C18 trapping microcolumn (5 mm by 300 μm, with an inner diameter of 5 μm; Agilent) in H2O-acetonitrile at 97:3 (vol/vol) plus 0.1% formic acid at a flow rate of 10 μl/min. After 5 min, they were back-flush eluted and separated on a reversed-phase nanocolumn, a ZORBAX 300SB C18 column (75 μm inner diameter by 15 cm, 3.5 μm; Agilent), at a flow rate of 300 nl/min with a gradient from 5 to 85% acetonitrile in 0.1% formic acid as follows: 5 min at 0% solvent B, from 0 to 25% solvent B in 35 min, 25 to 50% solvent B in 15 min, 50 to 90% solvent B in 5 min, 90% solvent B during 10 min, 90 to 0% solvent B in 5 min, and 0% for 15 min (total time, 90 min).
For spraying, a 400-nozzle ESI (electrospray ionization) chip (Advion Biosciences) was used with a voltage of 1.65 kV, and the mass spectrometer capillary transfer temperature was set at 200°C. In data-dependent acquisition controlled by Xcalibur 2.0.7 software (Thermo Fisher), the four most intense precursor ions detected in the full MS survey performed in the Orbitrap (range, 350 to 1,500 m/z; resolution, 60,000 at m/z 400) were selected and fragmented. MS-MS was triggered by a minimum signal threshold of 10,000 counts and carried out at a relative collision energy of 35%, with isolation width of 4.0 atomic mass units (amu). Only precursors with a charge higher than 1 were selected for collision-induced dissociation (CID) fragmentation and fragment ions were analyzed in the LTQ linear trap. The m/z of fragmented precursors was then dynamically excluded, with a tolerance of 0.01 amu, from any selection during 120 s.
From raw files, MS-MS spectra were exported as mgf (Mascot generic file, text format) files by using the extract_msn.exe script (Thermo Fisher) with the following settings: peptide mass range, 500 to 5,000; minimum total ion intensity threshold, 500; minimum number of fragment ions, 15; minimum signal-to-noise ratio needed for a peak to be written, 3.
Samples were analyzed using Mascot 2.2 (Matrix Science, London, United Kingdom). Mascot was set up to search a custom-built database containing the sequences of the vacuolar proteinase B (YscB) and of contaminants (enzymes and keratins). Semispecific cleavage at K and R (not before P) was used as the enzyme definition, with a maximum of one missed cleavage allowed. Mascot was searched with a fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 10 ppm. The iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine, and protein N acetylation were specified as variable modifications.
Phylogenetic analysis.
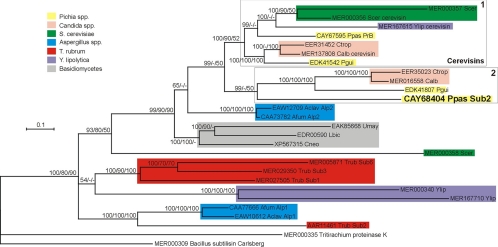
Amino acid sequences of P. pastoris PrB and Sub2 were used as a query to seek orthologous sequences in 54 available fungal genomes by using the blastp algorithm. A total of 27 subtilisins from selected species and showing a blast score of >200 were submitted to phylogenetic analyses. The subset included sequences from yeasts (from the subphylum Saccharomycotina, Saccharomyces cerevisiae, P. pastoris, Pichia guilliermondii, Candida albicans, Candida tropicalis, and Yarrowia lipolytica), filamentous ascomycetes (from the subphylum Pezizomycotina, Aspergillus fumigatus, Aspergillus clavatus, and Trichophyton rubrum), and basidiomycetes (Ustilago maydis, Laccaria bicolor, and Cryptococcus neoformans). Proteinase K from Tritirachium album was also included, and the type sequence of S8 Merops peptidase family, subtilisin Carlsberg from Bacillus licheniformis, was used as a potential outgroup.
Amino acid sequences were aligned using ClustalW as implemented in BioEdit Sequence Alignment Editor software (18). Phylogenetic analyses were performed using the following reconstruction methods and parameters: PhyML (17) with Shimodaira-Hasegawa (SH)-like approximate likelihood ratio test, four substitution rate categories, and estimation of gamma distribution parameter and proportion of invariable sites; BIONJ (12), using a Dayhoff point accepted mutation (PAM) substitution matrix; TNT (15) with sectorial search and tree fusing options; MrBayes (35) with general time-reversible (GTR) likelihood model and 100,000 Markov chain Monte Carlo generations. Phylogenetic trees were edited by using Dendroscope (20).
Nucleotide sequence accession number.
The SUB2 nucleotide sequence has been deposited in the GenBank database with the accession number GU46877.
RESULTS
Endogenous proteolytic activity of P. pastoris GS115 and KM71.
By use of SDS-PAGE gels containing gelatin as a substrate, two different proteolytic activities were detected in the supernatant of P. pastoris cultures in methanol medium. One protease showed a distinct band with an apparent molecular mass of 37 kDa (Fig. 1). A second activity was represented by a large proteolytic zone with an apparent molecular mass between 55 and 80 kDa. The 37-kDa protease was completely inhibited when the gel was incubated in the presence of 1 mM PMSF but not in the presence of 1,10-phenanthroline or EDTA, suggesting that it was a serine protease (data not shown). The 55- to 80-kDa protease was not inhibited by PMSF or the aforementioned chelating agents. Gelatinolytic activities of both strains were stronger at pH 7.0 than they were at pH 4.0 (data not shown).
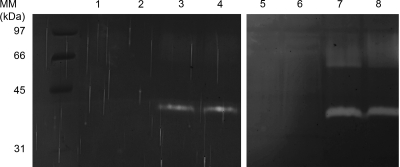
FIG. 1.
Native secreted subtilisin produced by P. pastoris GS115, KM71, and derivative strains in methanol medium. (Left panel) Ten microliters of P. pastoris culture supernatant was loaded onto the gel (lane 1, GS115-sub2; lane 2, KM71-sub2; lane 3, GS115; lane 4, KM71). (Right panel) P. pastoris GS115 and KM71 were grown to near saturation (OD of 20 at 600 nm) at 30°C in 10 ml of glycerol-based yeast medium for 2 days. Cells were harvested by centrifugation and resuspended in 2 ml of the same medium with 0.5% (vol/vol) methanol instead of glycerol and incubated for 2 days. For lanes 5 (GS115) and 6 (KM71), 10 μl of 50×-concentrated culture supernatant in glycerol-based yeast medium was loaded onto the gel. For lanes 7 (GS115) and 8 (KM71), 10 μl of 10×-concentrated culture supernatant in methanol medium as appropriate controls was loaded onto the gel. MM, molecular mass.
Cloning of a gene conferring native P. pastoris proteolytic activity.
Cloning experiments were performed before genome sequences were available (9, 27). In a first attempt, we tried to disrupt the gene encoding vacuolar protease B (PrB) in P. pastoris strain GS115. Protease B is a serine protease of the subtilisin family and was postulated to be secreted by P. pastoris (38). A gene encoding the putative P. pastoris PrB (PRB) was cloned from a P. pastoris genomic library as described in Materials and Methods. A central part of PRB was subsequently amplified using P1 and P2 as primers (Table 1 ) and using genomic DNA of P. pastoris GS115 as a template. The PCR product (785 bp) was digested by BamHI and XhoI restriction enzymes and ligated to the large fragment of P. pastoris expression vector pPICZA cut with BglII and XhoI. The resulting plasmid (pBL1263) corresponded to plasmid pPICZA in which the AOX1 promoter was replaced by part of PRB. Plasmid pBL1263 was cut by a unique BglII restriction site in the cloned fragment, and linearized DNA was used to transform P. pastoris GS115. Transformants resistant to Zeocin were screened for gelatinolytic activities on SDS-PAGE and verified for targeted integration of pBL1263 in GS115 by Southern blotting (3; data not shown). As a result, PRB-targeted disruption did not abolish the gelatinolytic activity in the P. pastoris cell culture supernatant. Therefore, we attempted to clone another gene encoding a subtilisin for subsequent disruption.
TABLE 1.
Primers used in this study
| Primera | Oligonucleotide sequence (5′-3′)b | Location | PCR product size (bp) (cloning site) |
|---|---|---|---|
| P1 | GTTGGATCCATCCAGCAGTTGCTTTCATTG | P. pastoris PRB | 785 (BamHI-XhoI) |
| P2 | CTTCTCGAGAGTACCAGACAAGGTAGCAG | Complement of PRB | |
| P3 | GGYCACGGXACXCACGTXGCXGGXAC | Subtilisin genes | 495 |
| P4 | GTGRGGXGTXGCCATXGAXGTXCC | Complement of subtilisin genes | |
| P5 | GTTTTCGAAGAAATGAAAATTATCAGATTAGCATCA | SUB2 | 1,455 (SfuI-XhoI) |
| P6 | CTTCTCGAGTTAATAATCCCAAAATTCAGT | Complement of SUB2 |
P3 and P4 were designed on the basis of alignments of nucleotide sequences encoding A. fumigatus Alp1 (GenBank CAA77666), Y. lipolytica secreted alkaline protease (MEROPS000340), S. cerevisiae PrB, C. albicans PrB (MEROPS137808), and C. albicans putative secreted subtilisin (MEROPS16558).
Restriction sites are indicated in italics.
Two primers, P3 and P4 (Table 1), were designed from an alignment of genes encoding known fungal subtilisins. PCR amplification of P. pastoris genomic DNA produced two 400-bp products which could be distinguished by BglII digestion. The two fragments were separately cloned into the multiple cloning site of the pDrive vector by using a PCR fragment cloning kit and sequenced. One fragment encoded part of PrB, and the second fragment apparently encoded an amino acid sequence from another subtilisin, which was called Sub2. The latter fragment was recovered from the pDrive construct after BamHI-XhoI digestion and ligated to the large fragment of P. pastoris expression vector pPICZA digested with restriction enzymes BglII and XhoI. The resulting plasmid (pKS1) cut by Bsu96I (SauI) at a unique restriction site was used to transform P. pastoris GS115 and KM71 (Fig. 2). Transformants resistant to Zeocin were screened on SDS-PAGE gels to which gelatin was added. One GS115 transformant out of 10 and 1 KM71 transformant out of 8 did not show any 37-kDa proteolytic component in the supernatant after growth in methanol medium (Fig. 1). These clones were designated GS115-sub2 and KM71-sub2, respectively.
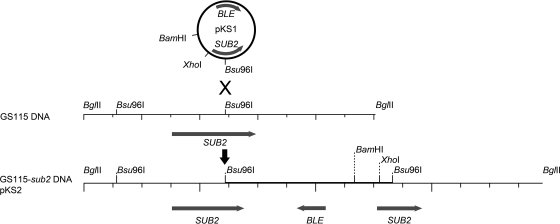
FIG. 2.
Chromosomal integration of pKS1 in GS115 and KM71 transformants. SUB2, P. pastoris SUB2 gene coding for the secreted subtilisin; BLE, Streptoalloteichus hindustanus BLE gene for resistance to Zeocin. The sequence of the 9-kb BglII-BglII fragment in GS115 and KM71 transformants is identical to the sequence of pKS2.
In order to clone the full-length SUB2 gene, 1-μg samples of GS115-sub2 genomic DNA were separately digested by different enzymes (DraI, BglII, ClaI, PstI, and EcoRI) and religated, and the products were then used to transform E. coli. Five hygromycin-resistant clones obtained from DNA digested by BglII were shown to harbor a 9-kb plasmid (pKS2; Fig. 2). Plasmid sequencing revealed the beginning and the end of SUB2 upstream and downstream of the pPICZA sequence, respectively (Fig. 2). The SUB2 sequence was identical to that encoding the protein with the GenBank accession number CAY68404 [“Vacuolar proteinase B (yscB), a serine protease of the subtilisin family”] in the Pichia pastoris chromosome 1 complete sequence (GenBank accession number FN3922319, locus tag PAS_chr1-4_548) (9).
Sub2 was predicted to be encoded by 1,431 nucleotides starting from the ATG codon and appeared to be synthesized as a precursor in a preprotein form of 477 amino acids. Inspection of the N-terminal amino acid sequence of the precursor beginning from the Met1 residue suggested the existence of a signal peptide in the protease precursor polypeptide with a hydrophobic core. Two putative signal peptidase cleavage sites in accordance with the −3 −1 von Heijne's rule (39) were found after the Ala and Ser residues at positions 16 and 18, respectively (Fig. 3). Based on alignment with different fungal secreted subtilisins (28), the N-terminal amino acid sequence of the mature Sub2 was apparently preceded by a precursor protein peptide (prepropeptide) of 168 amino acids, and the mature protein is 309 amino acids long, with a calculated molecular mass of 34 kDa and an isoelectric point at pH 5.69. In addition to Ser 402 (position relative to the Met1 residue of the prepropeptide), the alignment of Sub2 with other subtilisins revealed Asp and His residues of the catalytic triad in position 212 and 244, respectively.
FIG. 3.
Prepropeptide Sub2 amino acid sequence. Vertical arrows indicate putative signal sequence cleavage sites. The Asp, His, and Ser residues of the catalytic triad are indicated by asterisks. Detected MS peptides generated by tryptic digestion of Sub2 secreted by P. pastoris are underlined.
Biochemical characterization of Sub2 obtained from a Sub2-overproducing strain of P. pastoris.
DNA encoding Sub2 was amplified using primers P5 and P6 (Table 1) and P. pastoris genomic DNA as a target. The PCR product was digested with restriction enzymes SfuI and XhoI, for which a recognition site was previously designed at the 5′ extremity of the primers, and ligated to pPICZA cut with the same restriction enzymes. Linearized DNA of the generated plasmid was used to transform Pichia pastoris KM71, and Zeocin-resistant transformants were screened for Sub2 overproduction in the culture supernatant. One clone out of 10 was found to secrete Sub2 at a rate of 10 μg/ml. This clone was called KM71-SUB2 (Fig. 4).
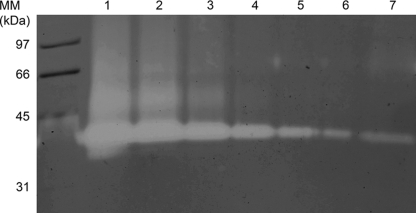
FIG. 4.
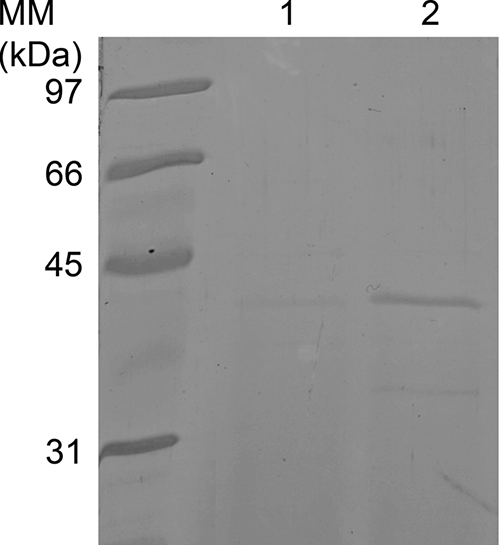
Overexpression of P. pastoris SUB2 in P. pastoris KM71 (lane 2). Lane 1 shows the culture supernatant of KM71 as a negative control. Ten microliters of P. pastoris culture supernatant was loaded onto the gel.
Sub2 produced by gene overexpression had the same electrophoretic mobility as the enzyme secreted by strain GS115 with an estimated mass of 37 kDa (Fig. 5). Identification of the heterologously produced Sub2 was also confirmed using ESI-LC-MS-MS on tryptic digestion of the detected 37-kDa protein in a gel slice (Fig. 3). MS analysis allowed the detection of several different peptides which matched from residues 169 to 468 but not from residues 1 to 168 of the prepropeptide Sub2 amino acid sequence. These results confirmed that cleavage of the prosequence occurred after residue 168 or few residues before, as it was suggested from alignment with other subtilisins.
FIG. 5.
Assessment of the amount of Sub2 secreted by P. pastoris GS115. In lanes 1 to 6, 10 ng, 3 ng, 1 ng, 300 pg, 100 pg, and 30 pg recombinant Sub2, respectively, were loaded on the gel. Lane 7, 10 μl of P. pastoris GS115 culture supernatant was loaded onto the gel.
Purification assays of recombinant Sub2 were attempted by ion-exchange chromatography. However, we observed that Sub2 underwent self-digestion at high concentrations. Therefore, further characterization of Sub2 was performed using enzyme from desalted culture supernatant. The enzyme was found to be active between pH 4.0 and 11.0, with a broad peak of optimum activity between pH 6.0 and 8.0 on resorufin-labeled casein and N-Suc-Ala-Ala-Pro-Phe-pNA. The specific activity of Sub2 and that of proteinase K used for comparison for N-Suc-Ala-Ala-Pro-Phe-pNA as a substrate at a pH of 7.0 were measured as 6.0 U and 26 mU/μg protein, respectively. Like other subtilisins (29), recombinant Sub2 was totally inhibited by PMSF, antipain, and chymostatin. Aprotinin and leupeptin inhibited the enzymatic activity by 15 to 20% at a concentration of 2 to 5 μg/ml. The enzyme was not inhibited by pepstatin, E64, bestatin, EDTA, and phosphoramidon.
Amount of Sub2 secreted by P. pastoris GS115.
Resorufin-labeled casein substrate as well as N-Suc-Ala-Ala-Pro-Phe-pNA was not sensitive enough to detect proteolytic activity in P. pastoris culture supernatants. Therefore, proteolytic activities of GS115 and KM71 culture supernatants were compared with serial dilutions of purified recombinant Sub2 extracts to assess the amount of protease secreted by the yeast (Fig. 5). One milliliter of culture supernatant was found to contain approximately 3 ng of Sub2.
SUB2 disruption in various P. pastoris strains producing secreted proteases and use of GS115-sub2 for heterologous protease production.
Pichia pastoris GS115-DPPIV and GS115-LAP2, producing AfuDppIV and AfuLap2, respectively, were previously constructed (2, 29). Both strains were transformed by pKS1 cut at its unique Bsu96I restriction site, and Zeocin-resistant transformants were screened for SUB2 disruption. One clone producing AfuDppIV and one clone producing AfuLap2 which no longer produced Sub2 were retained and called GS115-DPPIV-sub2 and GS115-LAP2-sub2, respectively (Fig. 6). Identical results were obtained by transformation of P. pastoris GS115-sub2 with previously used expression plasmids encoding recombinant AfuLap2 and AfuDppIV (2, 31).
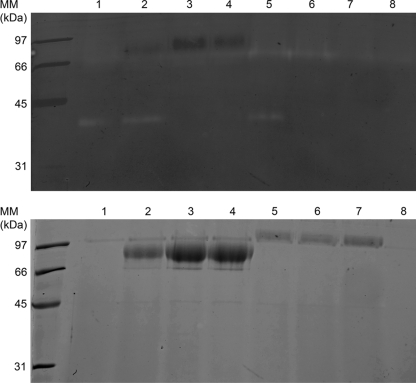
FIG. 6.
Use of GS115-sub2 for heterologous protease production (lanes 3 and 6) and SUB2 disruption in P. pastoris strains producing secreted proteases (lanes 4 and 7). Lane 1, GS115; lane 2, GS115-LAP2; lane 3, GS115-sub2 transformed with pKJ113-LAP2; lane 4, GS115-LAP2 transformed with pKS1 (SUB2 disruption in an AfuLap2-producing strain); lane 5, GS115-DPPIV; lane 6, GS115-sub2 transformed with pKJ113-DPPIV; lane 7, GS115-DPPIV transformed with pKS1 (SUB2 disruption in a DppIV-producing strain); lane 8, GS115-sub2. Ten microliters of P. pastoris GS115 culture supernatant was loaded onto the gel. The samples were analyzed by SDS-PAGE with a separation gel of 12% polyacrylamide stained with Coomassie blue (lower panel). To detect endoproteolytic activity of culture supernatant, gelatin at a concentration of 0.1% was added to the gel (upper panel).
Phylogenetic analyses.
Most selected yeast sequences branched into two derived phylogenetic groups (Fig. 7). Pichia pastoris PrB could be unambiguously identified as the orthologue of cerevisin proteases from S. cerevisiae, Y. lipolytica, C. albicans, and P. guilliermondii (clade 1; Fig. 7). Depending on the tree reconstruction methods used, P. pastoris Sub2 showed slight differences in branching. It formed a monophyletic group (clade 2; Fig. 7) together with hypothetical subtilisins from Candida spp. and P. guilliermondii in Bayesian (MrBayes) and distance (BIONJ) analyses, whereas it branched at the base of the group formed by clades 1 and 2 in maximum likelihood (PhyML) and parsimony (TNT) analyses. Clade 2 also comprised single Sub2 orthologues from other yeasts, namely, Debaryomyces hansenii, Lodderomyces elongisporus, Pichia stipitis, and Clavispora lusitaniae (data not shown). Pichia pastoris Sub2 showed 44 to 46% identity similarity with putative orthologues in other yeast species from clade 2. Both clades 1 and 2 are clearly yeast specific: vacuolar and secreted subtilisins from basidiomycetes and from filamentous ascomycetes (Aspergillus spp. and Trichophyton rubrum) formed the basal groups of the tree (Fig. 7).
FIG. 7.
Phylogenetic position of Pichia pastoris Sub2 (CAY68404) relative to other fungal proteases of the S8 family inferred from a Bayesian analysis of amino acid sequences. The numbers at nodes represent clade credibility or bootstrap values (100 replicates) obtained with MrBayes/PhyML/BIONJ methods, respectively. Nodes with credibility values lower than 50 were collapsed. Subtilisin Carlsberg from Bacillus licheniformis was used as a potential outgroup. Each sequence is identified by its GenBank or Merops accession number, species abbreviation, and protease name (if any). The colors of the rectangles correspond to specific species or taxonomic groups. Clade 1 comprises all cerevisin sequences; clade 2 comprises Pichia pastoris (Ppas) Sub2 and its putative orthologues from other yeast species. The scale bar indicates the number of expected changes per site. Scer, S. cerevisiae; Ylip, Y. lipolytica; Ctrop, C. tropicalis; Calb, Candida albicans; Pgui, P. guilliermondii; Aclav, A. clavatus; Afum, A. fumigatus; Umay, U. maydis; Lbic, Laccaria bicolor; Cneo, C. neoformans; Trub, T. rubrum.
DISCUSSION
The P. pastoris genome harbors two genes encoding serine proteases of the subtilisin family, PRB coding for the vacuolar proteinase B and SUB2 coding for the protease which gained particular attention in the present work. We have shown that Sub2 is secreted by P. pastoris in methanol medium in a high-cell-density process but not in the same medium containing glycerol as a carbon source instead of methanol (Fig. 1). While P. pastoris PrB is clearly orthologous to other vacuolar proteases (cerevisins) found in most yeast genomes, Sub2 belongs to another group of proteases which contains other putative yeast subtilisins (Fig. 7). Putative orthologues of Sub2 were found in only 7 out of 17 yeast genomes (subphylum Saccharomycotina) available. Pichia pastoris Sub2 is believed to be the first of these subtilisins identified in yeast culture supernatant. The derived phylogenetic positions of most yeast subtilisins compared to those of the basidiomycetes and of filamentous ascomycetes suggest a relatively recent radiation through gene duplication.
During the production of heterologous proteins in a high-cell-density process, some of the secreted heterologous proteins produced by P. pastoris were found to be subjected to substantial proteolysis (21, 22, 38). The major store of proteolytic activity in yeasts is located within the lumen of the vacuolar compartment (23), and previous reports disclosed that the proteolytic degradation of secreted recombinant proteins in P. pastoris was due to the release of proteases in the culture medium caused by cell lysis (27, 38). While growth at high cell density enables the production of heterologous proteins in remarkably high yields, it also increases the level of vacuolar proteases in the fermentation media (14). Using P. pastoris as an expression system to produce heterologous secreted protein, one should also be aware of the presence of secreted Sub2 in culture supernatant. The nature of the 55- to 80-kDa proteolytic activity was not elucidated, but similar 55- to 80-kDa activity was located in the periplasmic fraction of P. pastoris (38). It is possible that the protease is released in the culture supernatant either after cell lysis or by diffusion though the cell wall.
Apparently, Sub2 was insensitive to 55- to 80-kDa proteolytic activity. It is likely that an organism adapts to secrete proteins which are immune to degradation by its own proteases. For instance, in other fungi, such as Aspergillus spp. and dermatophytes, multiple proteases are concomitantly secreted in culture supernatant and are not degraded (13, 33). In addition, the acquisition of the correct three-dimensional structure may protect secreted proteins from proteolysis. However, various attempts to suppress proteolytic degradation of secreted recombinant proteins in the P. pastoris culture supernatant were undertaken. For example, conditions under which recombinant strains are grown were modified so as to inhibit protease activity. Adjusting the pH of the culture to 6.0 or lowering the temperature of the medium allowed a substantial reduction of proteolytic activity (21, 26). Determining the optimum induction time in methanol medium for recombinant protein production was also found to be important (22). To inhibit proteolysis in culture supernatant, vacuolar protease-deficient strains of P. pastoris were also constructed (14). Although vacuolar proteases are required for several developmental transitions in the life cycle of yeast cells (e.g., sporulation), they are dispensable for vegetative growth. The same is true for Sub2, as SUB2 disruption does not alter the viability of P. pastoris. Therefore, P. pastoris sub2-deficient strains could be used to suppress one of the causes of secreted heterologous enzyme proteolysis. Although secreted recombinant proteases were not subject to proteolysis, GS115-sub2 was used as a tool for the production of exoproteases without endogenous subtilisin contamination.
Acknowledgments
We thank Massimo Lurati for critical review of the manuscript and assistance with the English and Marina Fratti for helpful discussions and technical assistance.
This work was supported by the Swiss National Foundation for Scientific Research, grant 320030-1179641.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Beauvais, A., M. Monod, J. P. Debeaupuis, M. Diaquin, H. Kobayashi, and J. P. Latgé. 1997. Biochemical and antigenic characterization of a new dipeptidyl-peptidase isolated from Aspergillus fumigatus. J. Biol. Chem. 272:6238-6244. [DOI] [PubMed] [Google Scholar]
- 2.Beauvais, A., M. Monod, J. Wyniger, J. P. Debeaupuis, E. Grouzmann, N. Brakch, J. Svab, A. G. Hovanessian, and J. P. Latge. 1997. Dipeptidyl-peptidase IV secreted by Aspergillus fumigatus, a fungus pathogenic to humans. Infect. Immun. 65:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beggah, S., B. Léchenne, U. Reichard, S. Foundling, and M. Monod. 2000. Intra- and intermolecular events direct the propeptide-mediated maturation of the Candida albicans secreted aspartic proteinase Sap1p. Microbiology 146:2765-2773. [DOI] [PubMed] [Google Scholar]
- 4.Borelli, C., E. Ruge, J. H. Lee, M. Schaller, A. Vogelsang, M. Monod, H. C. Korting, R. Huber, and K. Maskos. 2008. X-ray structures of Sap1 and Sap5: structural comparison of the secreted aspartic proteinases from Candida albicans. Proteins 72:1308-1319. [DOI] [PubMed] [Google Scholar]
- 5.Borg-von Zepelin, M., S. Beggah, K. Boggian, D. Sanglard, and M. Monod. 1998. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28:543-554. [DOI] [PubMed] [Google Scholar]
- 6.Couderc, R., and J. Baratti. 1980. Oxidation of methanol by the yeast Pichia pastoris: purification and properties of alcohol oxidase. Agric. Biol. Chem. 44:2279-2289. [Google Scholar]
- 7.Cregg, J. M., K. R. Madden, K. J. Barringer, G. P. Thill, and C. A. Stillman. 1989. Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris. Mol. Cell. Biol. 9:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DelMar, E. G., C. Largman, J. W. Brodrick, and M. C. Geokas. 1979. A sensitive new substrate for chymotrypsin. Anal. Biochem. 99:316-320. [DOI] [PubMed] [Google Scholar]
- 9.De Schutter, K., Y. C. Lin, P. Tiels, A. Van Hecke, S. Glinka, J. Weber-Lehmann, P. Rouzé, Y. Van de Peer, and N. Callewaert. 2009. Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 6:561-566. [DOI] [PubMed] [Google Scholar]
- 10.De Viragh, P. A., D. Sanglard, G. Togni, R. Falchetto, and M. Monod. 1993. Cloning and sequencing of two Candida parapsilosis genes encoding acid proteases. J. Gen. Microbiol. 139:335-342. [DOI] [PubMed] [Google Scholar]
- 11.Ellis, S. B., P. F. Brust, P. J. Koutz, A. F. Waters, M. M. Harpold, and T. R. Gingeras. 1985. Isolation of alcohol oxidase and two other methanol regulatable genes from the yeast Pichia pastoris. Mol. Cell. Biol. 5:1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gascuel, O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685-695. [DOI] [PubMed] [Google Scholar]
- 13.Giddey, K., M. Monod, J. Barblan, A. Potts, P. Waridel, C. Zaugg, and M. Quadroni. 2007. Comprehensive analysis of proteins secreted by Trichophyton rubrum and Trichophyton violaceum under in vitro conditions. J. Proteome Res. 6:3081-3092. [DOI] [PubMed] [Google Scholar]
- 14.Gleeson, M. A., and B. D. Howard. November 1997. Genes which influence Pichia proteolytic activity, and uses therefor. U.S. patent 5,691,166.
- 15.Goloboff, P., J. Farris, and K. Nixon. 2008. TNT: a free program for phylogenetic analysis. Cladistics 24:774-786. [Google Scholar]
- 16.Grossberger, D. 1987. Minipreps of DNA from bacteriophage lambda. Nucleic Acids Res. 15:6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guindon, S., and O. Gascuel. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 18.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 19.Higgins, D. R., and J. M. Cregg. 1998. Pichia protocols. Humana Press, Totowa, NJ.
- 20.Huson, D. H., D. C. Richter, C. Rausch, T. Dezulian, M. Franz, and R. Rupp. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jahic, M., F. Wallberg, M. Bollok, P. Garcia, and S. O. Enfors. 2003. Temperature limited fed-batch technique for control of proteolysis in Pichia pastoris bioreactor cultures. Microb. Cell Fact. 18:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahic, M., M. Gustavsson, A. K. Jansen, M. Martinelle, and S. O. Enfors. 2003. Analysis and control of proteolysis of a fusion protein in Pichia pastoris fed-batch processes. J. Biotechnol. 102:45-53. [DOI] [PubMed] [Google Scholar]
- 23.Jones, E. W. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 194:428-453. [DOI] [PubMed] [Google Scholar]
- 24.Jousson, O., B. Léchenne, O. Bontems, B. Mignon, U. Reichard, J. Barblan, M. Quadroni, and M. Monod. 2004. Secreted subtilisin gene family in Trichophyton rubrum. Gene 339:79-88. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Li, Z., F. Xiong, Q. Lin, M. d'Anjou, A. J. Daugulis, D. S. Yang, and C. L. Hew. 2001. Low-temperature increases the yield of biologically active herring antifreeze protein in Pichia pastoris. Protein Expr. Purif. 21:438-445. [DOI] [PubMed] [Google Scholar]
- 27.Mattanovich, D., A. Graf, J. Stadlmann, M. Dragosits, A. Redl, M. Maurer, M. Kleinheinz, M. Sauer, F. Altmann, and B. Gasser. 2009. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb. Cell Fact. 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mignon, B., M. Swinnen, J. P. Bouchara, M. Hofinger, A. Nikkels, G. Pierard, C. Gerday, and B. Losson. 1998. Purification and characterization of a 31.5 kDa keratinolytic subtilisin-like serine protease from Microsporum canis and evidence of its secretion in naturally infected cats. Med. Mycol. 36:395-404. [PubMed] [Google Scholar]
- 29.Monod, M., G. Togni, L. Rahalison, and E. Frenk. 1991. Isolation and characterisation of an extracellular alkaline protease of Aspergillus fumigatus. J. Med. Microbiol. 35:23-28. [DOI] [PubMed] [Google Scholar]
- 30.Monod, M., G. Togni, B. Hube, and D. Sanglard. 1994. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol. Microbiol. 13:357-368. [DOI] [PubMed] [Google Scholar]
- 31.Monod, M., B. Léchenne, O. Jousson, D. Grand, C. Zaugg, R. Stocklin, and E. Grouzmann. 2005. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology 151:145-155. [DOI] [PubMed] [Google Scholar]
- 32.Monod, M. 2008. Secreted proteases from dermatophytes. Mycopathologia 166:285-294. [DOI] [PubMed] [Google Scholar]
- 33.Monod, M., O. Jousson, and U. Reichard. 2009. Aspergillus fumigatus secreted proteases, p. 87-106. In J.-P. Latgé and W. J. Steinbach (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC.
- 34.Reichard, U., B. Léchenne, A. R. Asif, F. Streit, E. Grouzmann, O. Jousson, and M. Monod. 2006. Sedolisins, as new class of secreted proteases from Aspergillus fumigatus with endoprotease or tripeptidyl-peptidase activity at acidic pH. Appl. Environ. Microbiol. 72:1739-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronquist, F., and J. P. Huelsenbeck. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 37.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 38.Sinha, J., B. A. Plantz, M. Inan, and M. M. Meagher. 2005. Causes of proteolytic degradation of secreted recombinant proteins produced in methylotrophic yeast Pichia pastoris: case study with recombinant ovine interferon-tau. Biotechnol. Bioeng. 89:102-112. [DOI] [PubMed] [Google Scholar]
- 39.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 41.Zaugg, C., O. Jousson, B. Léchenne, P. Staib, and M. Monod. 2008. Trichophyton rubrum secreted and membrane-associated carboxypeptidases. Int. J. Med. Microbiol. 298:669-682. [DOI] [PubMed] [Google Scholar]