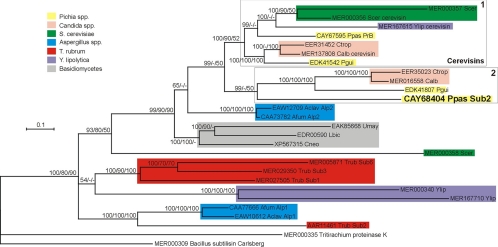
FIG. 7.
Phylogenetic position of Pichia pastoris Sub2 (CAY68404) relative to other fungal proteases of the S8 family inferred from a Bayesian analysis of amino acid sequences. The numbers at nodes represent clade credibility or bootstrap values (100 replicates) obtained with MrBayes/PhyML/BIONJ methods, respectively. Nodes with credibility values lower than 50 were collapsed. Subtilisin Carlsberg from Bacillus licheniformis was used as a potential outgroup. Each sequence is identified by its GenBank or Merops accession number, species abbreviation, and protease name (if any). The colors of the rectangles correspond to specific species or taxonomic groups. Clade 1 comprises all cerevisin sequences; clade 2 comprises Pichia pastoris (Ppas) Sub2 and its putative orthologues from other yeast species. The scale bar indicates the number of expected changes per site. Scer, S. cerevisiae; Ylip, Y. lipolytica; Ctrop, C. tropicalis; Calb, Candida albicans; Pgui, P. guilliermondii; Aclav, A. clavatus; Afum, A. fumigatus; Umay, U. maydis; Lbic, Laccaria bicolor; Cneo, C. neoformans; Trub, T. rubrum.