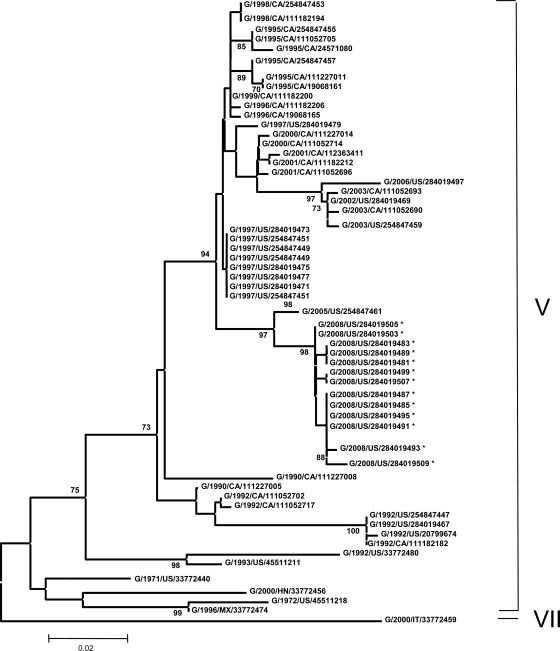
FIG. 4.
Phylogenetic analysis of 374 nucleotides corresponding to the partial sequence of the F gene of 68 NDV isolates. The evolutionary history was inferred using the neighbor-joining method (19). The optimal tree with the sum of the branch length of 0.60932040 is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (7). The tree is drawn to scale, with the branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method (13, 20) and are in the units of the number of base substitutions per site. The codon positions included were 1st, 2nd, 3rd, and noncoding positions. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 354 positions in the final data set. Phylogenetic analyses were conducted in the MEGA4 (20). Not all isolates included in the analysis are shown on the tree because they group with other isolates on the same position on the branch. Isolates G/2008/US/284019511, -284019514, -284019501, -284019517, and -284019520 group with G/2008/US/284019485, -284019487, -284019491, and -284019495. Isolate G/2008/US/284019523 groups with G/2008/US/284019499 and -284019507. Isolates G/2008/US/284019526 and -284019532 group with G/2008/US/284019481, -284019483, and -284019489. Isolate G/2008/US/284019529 groups with G/2008/US/284019503 and -284019505. *, isolates first characterized in this study.