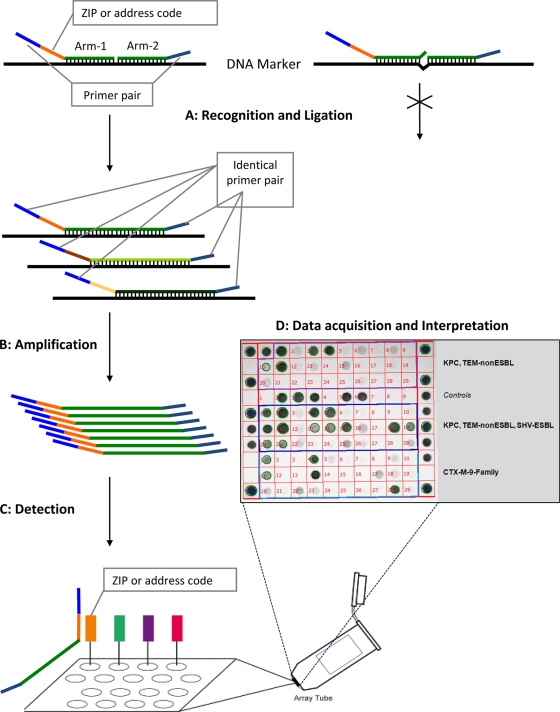
FIG. 1.
Schematic flowchart representing the different steps used by the Check-Points KPC/ESBL platform to recognize specific bla genes. (Step A) Target DNA recognition and ligation (thermocycling conditions, 95°C for 3 min, 24 cycles of 95°C for 30 s and 65°C for 5 min, and 98°C for 2 min [total, 2.75 h]). Each target-specific probe consists of two oligonucleotide probe arms that are used to detect single nucleotide polymorphisms (SNPs). These two probe arms are connected by the ligase, generating a single probe molecule only when they perfectly match the target sequence. Only connected probe arms produce the labeled amplification products detected in step C. Every target-specific probe is equipped with the same consensus primer pair necessary for step B and a unique “ZIP code” necessary for step C. (Step B) PCR amplification of the target DNA sequences (thermocycling conditions, 95°C for 10 min, 35 cycles of 95°C for 5 s, 55°C for 30 s, and 72°C for 30 s, and 98°C for 2 min [total, 1.5 h]). Using a common primer pair, target ligated sequence templates labeled with specific “ZIP codes” are multiplied. (Step C) Detection (requiring approximately 1 to 2 h of processing, depending on the number of samples). Amplification products are targeted to specific addresses on the microarray. This targeting is dependent on the specific “ZIP code.” (Step D) Immediate acquisition of images by scanning of the microarray using the array tube reader and immediate interpretation of the acquired pictures by the use of dedicated software.