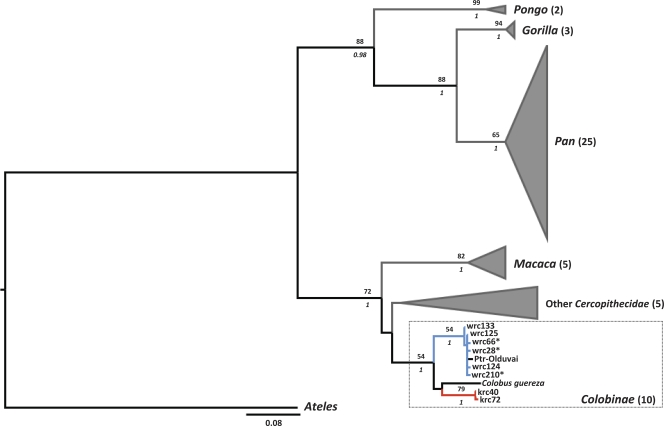
FIG. 3.
Maximum-likelihood tree based on the analysis of SFV partial pol sequences (252 bp). The topologies of Bayesian maximum clade credibility trees obtained under two different tree priors were similar. Branches leading to strains isolated from red colobus monkeys in Côte d'Ivoire are blue, those leading to strains isolated from Ugandan red colobus monkeys are red. Noncolobine tips are represented graphically to improve readability: in every case the number of strains represented is indicated between parentheses after family, subfamily or genus names. Numbers above branches represent bootstrap values (Bp), italicized numbers below branches represent posterior probability values (pp) obtained using the birth-death model. Bp and pp are indicated only where Bp is ≥50 and pp is ≥0.95. Asterisks indicate strains identified in the present study. Two of these strains were actually identified in more than one individual: wrc125 is identical to wrc3*, wrc12, wrc45*, wrc68*, wrc71*, wrc126, wrc127, wrc128, wrc129, wrc130, wrc131, wrc213*, wrc236*, and wrc276* (15 wrc sequences plus ptr-Leo); wrc133 is identical to wrc132*. Ptr, chimpanzee (P. troglodytes); krc, Eastern red colobus (P. r. tephrosceles); wrc, Western red colobus (P. b. badius).