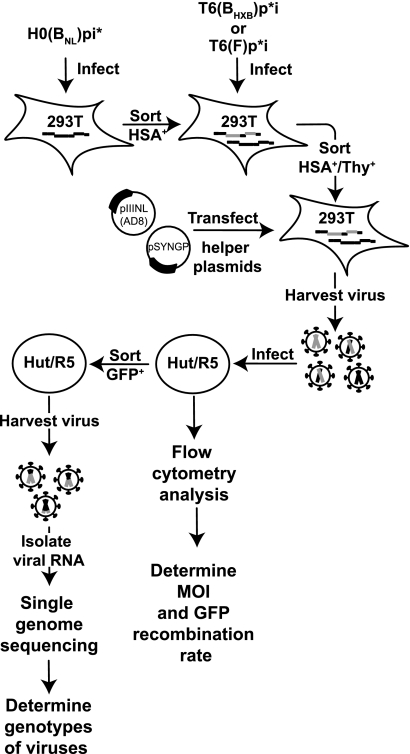
FIG. 2.
Protocol used to measure HIV-1 recombination in a single-cycle assay. Briefly, viruses containing the H0(BNL)pi* RNA were used to infect 293 cells at low MOI, and the resulting HSA+ cells were enriched by sorting. HSA+ cells were infected with viruses containing either T6(BHXB)p*i or T6(F)p*I at low MOI, and HSA+/Thy+ cells were enriched by cell sorting. Helper constructs pSYNGP and pIIINL(AD8), which expressed HIV-1 Gag/Gag-Pol and Env, respectively, were transfected into HSA+/Thy+ cell lines; viruses were harvested and used to infect fresh Hut/R5 cells. A portion of the infected Hut cells was analyzed by flow cytometry to determine the number of cells that expressed HSA, Thy, or GFP. These results were used to calculate the infection MOI and GFP recombination rate. Another portion of the infected cells was sorted to enrich GFP+ cells; viruses were harvested from GFP+ cells, and RNA was isolated and analyzed by single-genome sequencing to determine the genotypes of the viruses.