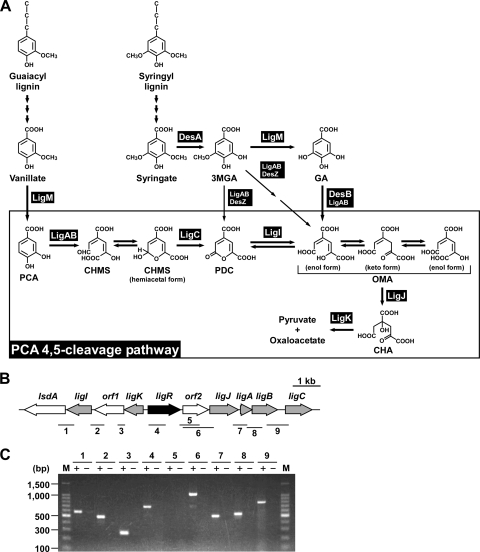
FIG. 1.
Catabolic pathway for vanillate and syringate and gene organization of the PCA45 pathway genes in Sphingobium sp. strain SYK-6. (A) Catabolism of vanillate and syringate. Enzymes: LigM, vanillate/3MGA O-demethylase; LigA and LigB, small and large subunits of PCA 4,5-dioxygenase; LigC, CHMS dehydrogenase; LigI, PDC hydrolase; LigJ, OMA hydratase; LigK, CHA aldolase/oxaloacetate decarboxylase; DesA, syringate O-demethylase; DesZ, 3MGA 3,4-dioxygenase; DesB, GA dioxygenase. Abbreviations: PCA, protocatechuate; CHMS, 4-carboxy-2-hydroxymuconate-6-semialdehyde; 3MGA, 3-O-methylgallate; GA, gallate; PDC, 2-pyrone-4,6-dicarboxylate; OMA, 4-oxalomesaconate; CHA, 4-carboxy-4-hydroxy-2-oxoadipate. (B) Organization of the PCA45 pathway genes. Regulatory genes, enzyme genes, and unidentified genes are indicated by black, gray, and white arrows, respectively. The bars and numbers below the genes indicate the locations of the amplified RT-PCR products shown in panel C. (C) RT-PCR analysis of the PCA45 pathway genes from SYK-6. Total RNA isolated from SYK-6 cells grown in the presence of PCA was used as a template. The sizes of molecular weight markers in lane M are indicated on the left. Lane numbers correspond to the numbers of the amplified regions indicated in panel B. “+” and “−“, with or without reverse transcriptase, respectively.