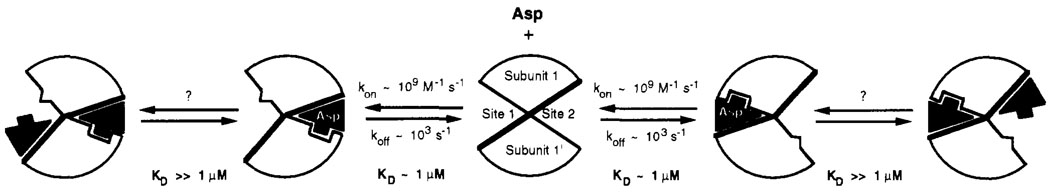
FIGURE 8.
Schematic model of the structural and kinetic aspects of aspartate binding. In the absence of bound aspartate, the ligand binding domain has, on average, a symmetrical structure possessing a C2 axis lying between the two identical subunits (center). The binding of the first aspartate molecule can occur at either of the two equivalent attractant binding sites, with the indicated association and dissociation rate constants. This binding event causes a conformational change within at least one subunit and destroys the symmetry of the dimer. The conformational change is communicated to the empty site, where the structure is altered such that the affinity for aspartate is significantly reduced (negative cooperativity). The binding of a second aspartate to this empty site was neither detected nor excluded by the present study; however, if it occurs it generates no detectable conformational change within the protein at the positions monitored by 19F NMR.