Abstract
Spo13 is a key meiosis-specific regulator required for centromere cohesion and coorientation, and for progression through two nuclear divisions. We previously reported that it causes a G2/M arrest and may delay the transition from late anaphase to G1, when overexpressed in mitosis. Yet its mechanism of action has remained elusive. Here we show that Spo13, which is phosphorylated and stabilized at G2/M in a Cdk/Clb-dependent manner, acts at two stages during mitotic cell division. Spo13 provokes a G2/M arrest that is reversible and largely independent of the Mad2 spindle checkpoint. Since mRNAs whose induction requires Cdc14 activation are reduced, we propose that its anaphase delay results from inhibition of Cdc14 function. Indeed, the Spo13-induced anaphase delay correlates with Cdc14 phosphatase retention in the nucleolus and with cyclin B accumulation, which both impede anaphase exit. At the onset of arrest, Spo13 is primarily associated with the nucleolus, where Cdc14 accumulates. Significantly, overexpression of separase (Esp1), which promotes G2/M and anaphase progression, suppresses Spo13 effects in mitosis, arguing that Spo13 acts upstream or parallel to Esp1. Given that Spo13 overexpression reduces Pds1 and cyclin B degradation, our findings are consistent with a role for Spo13 in regulating APC, which controls both G2/M and anaphase. Similar effects of Spo13 during meiotic MI may prevent cell cycle exit and initiation of DNA replication prior to MII, thereby ensuring two successive chromosome segregation events without an intervening S phase.
SPO13, a gene expressed uniquely in meiosis (Buckingham et al. 1990), was identified in a budding yeast strain found in nature that generated two-spored asci (Klapholz and Esposito 1980a). Subsequent analysis showed that it is required for faithful centromere segregation at MI (Klapholz and Esposito 1980b; Rutkowski and Esposito 2000), protection of Rec8 cleavage by separase during MI (Klein et al. 1999; Shonn et al. 2002), the recruitment of monopolin to centromeres (Toth et al. 2000; Rabitsch et al. 2003; Katis et al. 2004; Lee et al. 2004), and the establishment of two successive chromosome segregation events in wild-type cells (Klapholz and Esposito 1980b; Wagstaff et al. 1982; Rutkowski and Esposito 2000; Shonn et al. 2000). Although the action of Spo13 in centromere cohesion and coorientation has been studied extensively, the mechanism by which it regulates the timing and progression of meiotic cell divisions is unclear.
In earlier overexpression studies, it was proposed that Spo13 acts as a negative regulator of cell division, based on the finding that it caused a G2/M arrest in mitosis, when overexpressed from a heterologous promoter. In meiosis, Spo13 was thought to act transiently at two stages: first at G2/M to delay meiotic segregation until the machinery for reductional segregation is assembled, and second, at anaphase/mitotic exit (A/exit) to allow a second round of segregation without an intervening S phase (McCarroll and Esposito 1994). This hypothesis was based on the observation that SPO13 overexpression during sporulation delayed completion of MI and that the capacity for two consecutive divisions in a spo13 deletion mutant could be partially restored by slowing down meiosis. Moreover, SPO13 overexpression during meiosis suppressed the failure of cdc28-1 cells to execute MII at semi-permissive temperature, a defect that generates dyads with two diploid MI spores (Shuster and Byers 1989). Instead, mostly four-spored tetrads appeared, suggesting that Spo13 compensates for reduced CDK activity sufficiently to promote entry into a second M phase (MII) after MI, without passage through G1/S (McCarroll and Esposito 1994).
Progression through mitotic M phase and through the two meiotic divisions requires the activation of Cdc14 phosphatase. This coincides with the release of Cdc14 from its nucleolar inhibitor Net1 (Shou et al. 1999; Visintin et al. 1999) upon Net1 phosphorylation (Azzam et al. 2004; reviewed in Stegmeier and Amon 2004; Toth et al. 2007). Two different pathways positively regulate this process: the FEAR pathway (Cdc fourteen early anaphase release), which is active during the mitotic M phase, as well as the meiotic MI phase (Stegmeier et al. 2002; Marston et al. 2003), and the MEN (mitotic exit network (Geymonat et al. 2002; Stegmeier and Amon 2004)). Separase, a protease encoded by ESP1, is part of the FEAR pathway (Stegmeier et al. 2002), which couples cohesin cleavage with the early release of Cdc14 from the nucleolus (Queralt et al. 2006), thus promoting the G2/M transition. Separase is also implicated in MEN pathway activation and the promotion of the A/exit transition (Queralt et al. 2006).
Here we investigate how Spo13 overexpression provokes mitotic arrest. We exploited Spo13's behavior during mitosis to study its mechanism of action, since the ability to synchronize mitotic division, coupled with extensive knowledge of metaphase, anaphase, and mitotic exit events, allows questions to be addressed that are difficult to examine in meiosis. While overexpression studies can result in phenotypes that do not reflect normal activity, they have also provided important insights into gene function. Finally, we believe that the findings described for Spo13-dependent cell cycle arrest in mitotic cells may shed light on its normal meiotic function, since several hundred PCR-generated mutants in the SPO13 open reading frame selected for being deficient in mitotic arrest were also deficient for meiotic division (J. D. Fackenthal and R. E. Esposito, unpublished observation). This suggests that the meiotic and mitotic functions of Spo13 are closely related, if not inseparable.
MATERIALS AND METHODS
Strains and growth conditions:
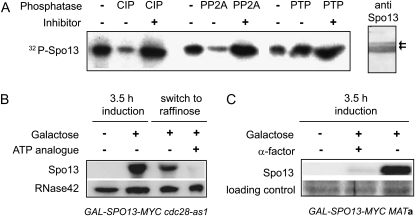
Table 1 lists the yeast strains used in this work. Most studies used isogenic homozygous diploids containing SPO13 or spo13fs fused to the GAL1 promoter (McCarroll and Esposito 1994) and integrated into the genome of W303 strains at the URA3 locus (this study). The native SPO13 gene is not expressed in mitosis and was therefore not deleted in these strains. Other tagged and deletion strains were constructed using standard transformation and PCR-based techniques (Longtine et al. 1998). Plasmids and overexpression constructs used are described in previous studies (Tinker-Kulberg and Morgan 1999; Bystricky et al. 2005). Standard growth and sporulation media were used (Williams et al. 2002). To induce Spo13 overexpression, cells were pregrown to mid/late log phase in YP with 3% raffinose. Galactose (2%) was then added and cells were sampled at various times after induction. Where indicated, nocodazole was added to cultures adjusted to 1% DMSO at a final concentration of 8 μg/ml. In vivo phosphorylation was carried out with a 15-min pulse of phosphate 32Pi (0.25 mm; 108 cpm/μmol phosphate). Spo13 was immunoprecipitated and analyzed by SDS-PAGE. Calf intestinal phosphatase (CIP), protein phosphatase 2A (PP2A), a Ser/Thr phosphatase, and protein tyrosine phosphatase (PTP), with or without the phosphatase inhibitors (0.1 m Na2HPO4, 20 μm okadaic acid, or 200 μm Na-vanadate) were used to assess phosphorylation.
TABLE 1.
Yeast strains
| Strain name | Relevant genotype (W303 background) |
|---|---|
| GA-583 | MATa/MATα GAL-spo13fs-URA3/GAL-spo13fs-URA3 MAD2∷mad2-KanMX/MAD2∷mad2-KanMX |
| GA-662 | MATa/MATα GAL-SPO13-URA3/GAL-SPO13-URA3 MAD2∷mad2-KanMX/MAD2∷mad2-KanMX |
| GA-4318 | MATa/MATα GAL-spo13fs-URA3/GAL-spo13fs-URA3 |
| GA-3419 | MATa/MATα GAL-SPO13-URA3/GAL-SPO13-URA3 |
| GA-4175 | MATa/MATα GAL-SPO13-GFP-KanMX/GAL-SPO13-KanMX pNOP1-CFP-HIS3 |
| GA-4355 | MATacdc28∷cdc28-as1-URA3 GAL-SPO13-13Myc-URA3 |
| GA-4388 | MATa/MATα GAL-spo13fs-URA3/GAL-spo13fs-URA3 cdc14∷CDC14-5GFP-TRP1/cdc14∷CDC14-5GFP-TRP1 pNOP1-CFP-HIS3 |
| GA-4390 | MATa/MATα GAL-SPO13-URA3/GAL-SPO13-URA3 cdc14∷CDC14-5GFP-TRP1/cdc14∷CDC14-5GFP-TRP1 pNOP1-CFP-HIS3 |
| GA-4393 | MATa/MATα GAL-spo13fs-URA3/GAL-spo13fs-URA3 cdc14∷CDC14-6HA-TRP1/cdc14∷CDC14-6HA-TRP1 |
| GA-4394 | MATa/MATα GAL-SPO13-URA3/GAL-SPO13-URA3 cdc14∷CDC14-6HA-TRP1/cdc14∷CDC14-6HA-TRP1 |
| GA-5230 | MATα GAL-SPO13 GAL-ESP1 cdc14∷CDC14-6HA-TRP1 |
| GA-5406 | MATaGAL-ESP1 cdc14∷CDC14-6HA-TRP1 |
| GA-5441 | MATaGAL-SPO13 cdc14∷CDC14-6HA-TRP1 |
| GA-5826 | MATaGAL-spo13fs-URA3 pds1∷PDS1-18MYC-TRP1 |
| GA-5829 | MATα GAL-SPO13-URA3 pds1∷PDS1-18MYC-TRP1 |
All strains were constructed for this study.
Western blot analysis:
Western blot analysis was performed with HRP-conjugated secondary antibodies. The signal was acquired using Quantity One software (Bio-Rad). Antibody dilutions were as follows: anti-Spo13 1/5000, anti-Clb2 1/2000 (Santa Cruz, SC 9071), anti-Sic1 1/100 (Santa Cruz SC 6712), and anti-tubulin (TAT-1) 1/2000. Antibodies against TrpE-Spo13 and GST-Spo13 fusions were used for Western blot analyses of the immunoprecipitated protein.
Immunofluorescence and live imaging:
Cells were grown as described above and fixed with a final concentration of 4% paraformaldehyde (PFA) for 20 min at room temperature. PFA was removed by washing with YPD at room temperature and cells were spheroplasted with lyticase and zymolyase. Cells were spotted onto a microscope slide, permeabilized, and processed for immunodetection as previously described (Gotta et al. 1996). Live imaging was performed using exponentially growing cultures at a density of 5–8 × 106 cells/ml. Cells were bound to concanavalinA-coated coverslips in a Ludin chamber (Life Imaging Services) and imaged in synthetic complete (SC) media at 30° using a Zeiss LSM510 confocal microscope. Step size for focal stacks was 200 nm. Imaging was performed as previously described (Varela et al. 2009).
Total RNA isolation, cRNA target synthesis, and GeneChip hybridization:
Culture conditions and array hybridization protocols were carried out as published, with a few modifications (Hochwagen et al. 2005). cRNA target molecules were prepared from 50-ml cultures at 3–5 × 107 cells/ml. Samples were hybridized to yeast S98 GeneChips (Affymetrix). Quality of total RNA and cRNA was monitored using RNA Nano 6000 chips processed using the 2100 BioAnalyzer (Agilent). A 220-μl hybridization cocktail containing heat-fragmented and biotin-labeled cRNA at a concentration of 0.05 μg/μl was injected into GeneChips and incubated at 45° on a rotator in a Hybridization Oven 640 (Affymetrix) overnight at 60 rpm. The arrays were washed and stained with a streptavidin-phycoerythrin conjugate (SAPE; Molecular Probes). The Gene Chips were processed in a GeneArray Scanner (Agilent) using the default settings.
Microarray data processing and statistical analysis:
CEL files containing the raw data were computed from DAT array image files using the statistical algorithm implemented in MAS 5.0 (Affymetrix). Data processing and quality control were done as previously described (Schlecht et al. 2008). First, probe sets for which we observed a signal ≥100 (empirical conservative background threshold) in at least one condition were identified. From these probe sets, the genes differentially expressed were identified by ANOVA. Genes were filtered using a P-value <0.005 and a standard deviation >0.3. Differentially expressed genes were then clustered using the k-means algorithm. Gene ontology was established by searching expression clusters for enrichment of functions using annotation, mapping, expression, and network (AMEN) analysis tool; see SourceForge at http://sourceforge.net/projects/amen), a microarray data analysis and visualization software suite (Chalmel and Primig 2008). For MIAME compliance, CEL feature-level data files are available via the EBI ArrayExpress public data repository at http://www.ebi.ac.uk/arrayexpress under accession no. E-MEXP-1459. The data are also accessible via the GermOnline database at http://www.germonline.org (Gattiker et al. 2007).
RESULTS
Inhibition of mitotic nuclear division by SPO13 is reversible and largely independent of Mad2:
Spo13 has been proposed to act as a cell-cycle regulator during meiosis (McCarroll and Esposito 1994). Loss-of-function mutants undergo a single division generating dyads with mostly equational sister–centromere segregation. Tetrad formation can be restored by slowing down meiosis with low temperatures or adding hydroxyurea (HU) to the medium (McCarroll and Esposito 1994). One model to explain this phenotype proposes that wild-type Spo13 acts as a transient negative regulator that slows down entry into and/or exit from M phase, until centromere cohesion, coorientation, and segregation occur, allowing the MI and MII divisions to proceed properly. One might expect, therefore, that overexpression of Spo13 from a heterologous promoter would provoke a gain-of-function phenotype in mitosis, such as a G2/M cell-cycle arrest. Indeed, this was found to be the case (McCarroll and Esposito 1994; Lee et al. 2002).
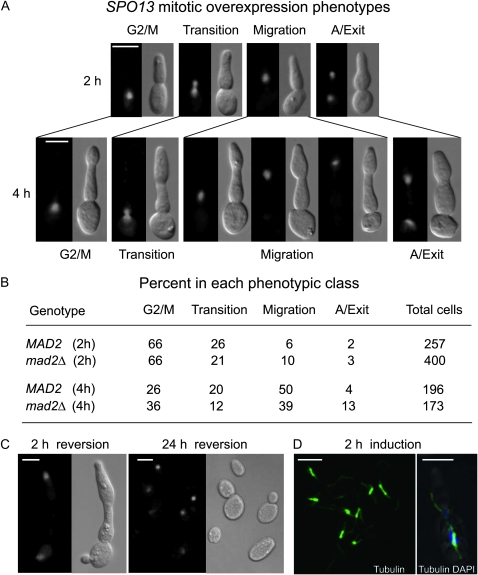
To examine more closely how overexpressed Spo13 affects the mitotic cell cycle, we created isogenic homozygous diploid cells bearing two integrated copies of wild-type SPO13 or a frame-shift mutation (spo13fs) under control of the inducible GAL1-10 promoter (hereafter called GAL-SPO13 or GAL-spo13fs). Log-phase cultures growing in raffinose were induced for 2 or 4 hr with galactose and stained with 4′,6-diamidino-2′-phenylindol-dihydrochloride (DAPI). Using fluorescence imaging, we observed four distinct classes of arrest after 2 hr on galactose, in cells expressing the wild-type but not the mutant gene (see supporting information, Figure S1). In class 1, the nucleus was present as a single body in the mother cell (G2/M phenotype); in class 2, the nucleus extended between mother and daughter cells (“transition” state); in class 3, a single nucleus migrated into the daughter cell (“migration”); in class 4, an elongated nucleus extended from mother to daughter cell, typical of anaphase (Figure 1A). It should be emphasized that the transition nuclei represent an ambiguous class, which could either reflect G2/M arrested cells or G2/M delayed cells moving toward nuclear migration. It is unlikely to reflect nuclei caught in the act of division since cell number remained constant during Spo13 overexpression. The appearance of long spindles in class 4 indeed indicates that these cells are arrested in anaphase (see below).
Figure 1.—
SPO13 overexpression phenotypes during mitosis. (A) Micrographs of DAPI staining and DIC images showing typical SPO13 overexpression phenotypes after 2 hr (upper) or 4 hr (lower) on galactose. (B) Quantification of the mitotic arrest phenotypes in GAL-SPO13 MAD2 (GA-3419) or GAL-SPO13 mad2 (GA-662) strains after 2 or 4 hr on galactose. The total number of cells presented for each strain stems from two independent experiments with identical phenotypic distributions. (C) Micrographs of DAPI staining and DIC images of GAL-SPO13 (GA-3419) showing the phenotypes 2 and 24 hr after transcriptional repression of GAL-SPO13 by return to raffinose-containing media (see materials and methods). (D) Immunofluorescence (IF) with anti-tubulin antibodies and DAPI staining of GAL-SPO13 (GA-3419) cells show misoriented spindles after 2 hr on galactose. Bar, 5 μm.
Similar nuclear phenotypes were detected at the 4-hr-arrest time point, although in different proportions. The number of G2/M arrested cells decreased from 66 to 26%, while class 3 cells with undivided migrating nuclei increased proportionately from 6 to 50% (Figure 1B). These findings suggest that a significant fraction of nuclei in the arrested cells eventually migrate into daughter cells. In contrast, the fraction of transition (class 2) and of anaphase (class 4) phenotypes remained essentially the same at 2 hr and 4 hr, suggesting that these classes might reflect cells arrested at two distinct points in the cell cycle: just prior to M phase, and in late anaphase, prior to cytokinesis. It should be noted that if class 2 represented G2/M arrested cells (class 1) moving toward migration (class 3), as considered earlier, then class 2 might be expected to decrease with time as migration increases, which does not occur.
Significantly, cell growth continued in all classes even though cytokinesis was inhibited, resulting in a pseudohyphal or hyperpolarized growth morphology (Figure 1A). Given that the arrest was not observed in cells overexpressing the spo13fs gene, we conclude that these phenotypes result from full-length Spo13 expression and not from growth on galactose (McCarroll and Esposito 1994). Subsequent immunofluorescence analysis for tubulin confirmed the predicted cell-cycle arrest points, by revealing a vast predominance of short, misoriented spindles (∼2.5 μm) with elongated cytoplasmic microtubules, even by 2 hr on galactose (Figure 1D). Only a small number of anaphase spindles were detected (Figures 4C and 7A).
Figure 4.—
Cdc14 colocalization with Nop1 in cells expressing SPO13 or spo13fs. (A) Cells expressing GAL-SPO13 CDC14-GFP pNOP1-CFP (GA-4390) or GAL-spo13fs CDC14-GFP pNOP1-CFP (GA-4388), as indicated, were visualized after 2 hr of growth on galactose. Selected cells arrested at G2/M (a) or in anaphase (b and c) and G1 (d) are enlarged in the middle and right panels. Nop1 is shown in red, Cdc14 in green, and colocalization of the two is shown in white (coloc). (B) Immunofluorescence for Cdc14-HA and anti-Nop1 on cultures expressing either GAL-SPO13 CDC14-6HA (GA-4394) or GAL-spo13fs CDC14-6HA (GA-4393) after 2 hr of induction with galactose. Micrographs show the colocalization of Cdc14 with Nop1 in red and their coincidence with DAPI staining (blue). In all categories of Spo13-induced arrest we see colocalization of Cdc14 and Nop1 staining, whereas in spo13fs-expressing cells we see colocalization in interphase but not in anaphase cells. (C) Immunofluorescence for Cdc14-HA or anti-tubulin, each combined with anti-Nop1, in a culture expressing GAL-SPO13 CDC14-6HA (GA-4394) after 2 hr of induction with galactose. (Top) Cdc14, Nop1, and DAPI are shown in yellow, green, and blue, respectively. Colocalization of Cdc14 and Nop1 is shown in red. (Bottom) anti-tubulin in green, anti-Nop1 in yellow, DAPI stain in blue. Bar, 5 μm. The diagram shows the distribution of proteins along the extended segregating chromosomes and microtubules (green) as in the micrographs.
Figure 7.—
Securin (Pds1) dynamics in the presence and absence of Spo13. Strains GA-5829 (GAL-SPO13 PDS1-18Myc) and GA5826 (GAL-spo13fs PDS1-18Myc) were grown on YP+raffinose to log phase and then galactose was added to induce expression of SPO13 or spo13fs. Both cultures were analyzed by immunofluorescence at 4 hr after induction. (B) The same strains described in A were arrested at the G2/M transition with nocodazole, then nocodazole was washed out with YP raffinose and cells were released into YP galactose. Samples were taken at the indicated time point and analyzed by Western blot. Antibodies against the indicated proteins were used. Bar, 5 μm.
To investigate whether the G2/M arrest is mediated by and dependent on the spindle checkpoint, we generated mad2 deletion strains in the GAL-SPO13 background. Again, SPO13 was overexpressed on galactose and the phenotypes were scored after 2 hr and 4 hr of induction. No substantial differences between the mad2 and MAD2 strains were found by 2 hr, suggesting that the initial arrest phenotype is largely independent of the spindle checkpoint (Figure 1B), and therefore are not due to impaired kinetochore function. By 4 hr, we note a threefold increase in anaphase cells in the mad2 mutant, possibly indicating a role for Mad2 in blocking progression from G2/M into a second arrest point in anaphase.
We further showed that the arrest due to SPO13 overexpression is reversible and that arrested cells can efficiently resume growth upon repression of the GAL1 promoter. Specifically, after 4 hr on galactose, SPO13-expressing cells were washed twice in glucose and then cultured in raffinose to repress GAL-SPO13 expression. After just 2 hr on raffinose, cells started to rebud. By 24 hr, cell morphology was identical to that observed before SPO13 induction (Figure 1C). Coincident with the reversal of arrest, Spo13 protein levels decreased significantly (see below and Figure 5A). These results are consistent with earlier observations of rapid SPO13 transcript turnover during transcriptional repression (Wang et al. 1987; Surosky and Esposito 1992), and the instability of Spo13 protein in meiosis once transcription ceases after MI (Katis et al. 2004; Sullivan and Morgan et al. 2007; J. D. Fackenthal and R. E. Esposito, unpublished results).
Figure 5.—
Western blot analysis of cell-cycle proteins. (A) Western blot analysis of samples at the indicated time points during galactose induction and after repression of the GAL promoter on raffinose media of GAL-SPO13 (GA-3419). Antibodies used are indicated at the right. (B) As in A, with strain GA-4318 expressing GAL-spo13fs.
On the basis of these initial results, we conclude that Spo13-dependent mitotic arrest prevents cell-cycle progression but not growth, stalling cells both at G2/M as well as in late mitosis in a reversible manner. Nuclei are nonetheless able to migrate into resulting pseudohyphae. The frameshift and reversibility controls argue that cell-cycle arrest stems directly from the high levels of Spo13 protein.
Spo13 is phosphorylated in vivo and depends on Cdc28/Clb for stability:
Immunoprecipitation of Spo13 from cells arrested in mitosis yielded multiple forms, which migrated more slowly than the major Spo13 band (Figure 2A, anti-Spo13). This suggested that Spo13 might be post-translationally modified. Because cdc28 mutant cells overexpressing SPO13 break through the mitotic arrest (McCarroll and Esposito 1994), we reasoned that Spo13 might be a target of a Cdk kinase and/or that the Cdc28/Clb complex might control Spo13 stability. Indeed, Spo13 contains 10 Cdk target motifs (SP/TP), as well as the conserved cyclin recognition site RLLEL, which enhances phosphorylation by cyclin/Cdk complexes when present on substrates (Brown et al. 1999).
Figure 2.—
Analysis of SPO13 phosphorylation and stability. (A) 32P-labeled band comigrates with Spo13-Myc, which forms multiple bands on a Western blot with anti-Myc antibody (right lane, anti-Spo13, see arrows) after immunoprecipitation. The 32P-labeled precipitates were incubated in the presence or absence of the indicated phosphatases (CIP, calf intestinal phosphatase; PP2A, protein phosphatase 2A; or PTP, protein tyrosine phosphatase) with or without an excess of the appropriate phosphatase inhibitors, Na2HPO4, okadaic acid, or Na-vanadate, as indicated. (B) Western blot analysis with anti-Myc antibody on strain GAL-SPO13-13MYC cdc28-as1 (GA-4355). Spo13-Myc was expressed for 3.5 hr and then repressed by washing twice in glucose. The culture was divided and further grown on raffinose for 3.5 hr. Where indicated, the ATP analog 1NM-PP1 was added to inactivate the cdc28-as1 allele. RNase42 indicates an abundant cytoplasmic RNA helicase used as a Western blot control. (C) Western blot analysis with anti-Myc antibody on GAL-SPO13-13MYC cdc28-as1 (GA-4355) in which SPO13-Myc expression was induced with galactose in the presence or absence of the yeast pheromone α-factor, which arrests cells in G1 phase. The loading control is an unidentified protein detected by Ponceau staining of the Western blot transfer.
To test this hypothesis, Myc-tagged SPO13 was expressed under control of the GAL1 promoter in the presence of 32P-labeled phosphate. Spo13-Myc was selectively immunoprecipitated and a 32P-labeled protein comigrating with Spo13 was recovered in the precipitate (Figure 2A). The incorporated label in the immunoprecipitated complex was sensitive to CIP and PP2A, but not PTP, suggesting that the protein does indeed carry Ser/Thr phosphorylation (Figure 2A).
To test whether Cdc28 was required for Spo13 stability, we generated a strain carrying GAL-SPO13-Myc and cdc28-as1, an allele of the unique Cdk in yeast that allows selective inhibition of Cdc28 by the addition of a small molecule inhibitor, 1NM-PP1 (Bishop et al. 2000). After induction of SPO13, the cells were switched to YPD media to repress SPO13 expression, and half the culture was incubated for 3.5 hr in the presence of 1NM-PP1 (10 μm final concentration), to inhibit Cdc28. Compared to the half that was exposed to solvent only, the culture incubated in 1NM-PP1 showed a strongly reduced Spo13-Myc signal, suggesting that Spo13 stability requires Cdc28 activity (Figure 2B). Consistent with this observation, we noted that Spo13-GFP levels were extremely low in G1-phase cells monitored by quantitative live microscopy (data not shown), and that in GAL-SPO13-Myc cells arrested in G1 by α-factor growth on galactose did not lead to Spo13-Myc accumulation (Figure 2C). Since Cdc28/Clb is absent in G1, we conclude that Cdc28/Clb kinase activity either directly or indirectly promotes Spo13 stability. These results are compatible with an independent study proposing that Spo13 is a target of the anaphase promoting complex (APC)-mediated degradation pathway in G1-phase cells (Sullivan and Morgan 2007).
Spo13 overexpression leads to reduced transcript levels of Ace2- and Swi5-dependent cell cycle genes:
While there is no evidence that Spo13 is or interacts with a transcription factor, we reasoned that genome-wide mRNA profiling analysis during SPO13 overexpression could reveal indirect effects that might shed light on why Spo13 provokes a mitotic arrest. To investigate this, we used high-density oligonucleotide microarrays (S98 GeneChips; Affymetrix bearing probes for 6430 annotated, protein-coding genes) and mRNA isolated from cells expressing either the GAL-SPO13 or GAL-spo13fs allele (Figure S1).
Independent cultures grown in duplicate on YP+raffinose were subjected to galactose induction of Spo13. Total RNA was isolated from samples at 0, 2, and 4 hr after the addition of galactose. RNA Nano6000 Chips and the 2100 BioAnalyzer (Agilent) were used to control the quality and homogeneity of total RNA and cRNA samples (Figure S2, A and B). Log2-transformed signals compared across the sample set using density and box plots were very similar and within the expected range, confirming reproducibility. Because global expression signals were similar at 0 and 2 hr, but distinct after 4 hr of Spo13 induction (in contrast to the frameshift control), at least some of the transcriptional changes appeared to be a consequence of the Spo13-induced cell-cycle arrest (Figure S3).
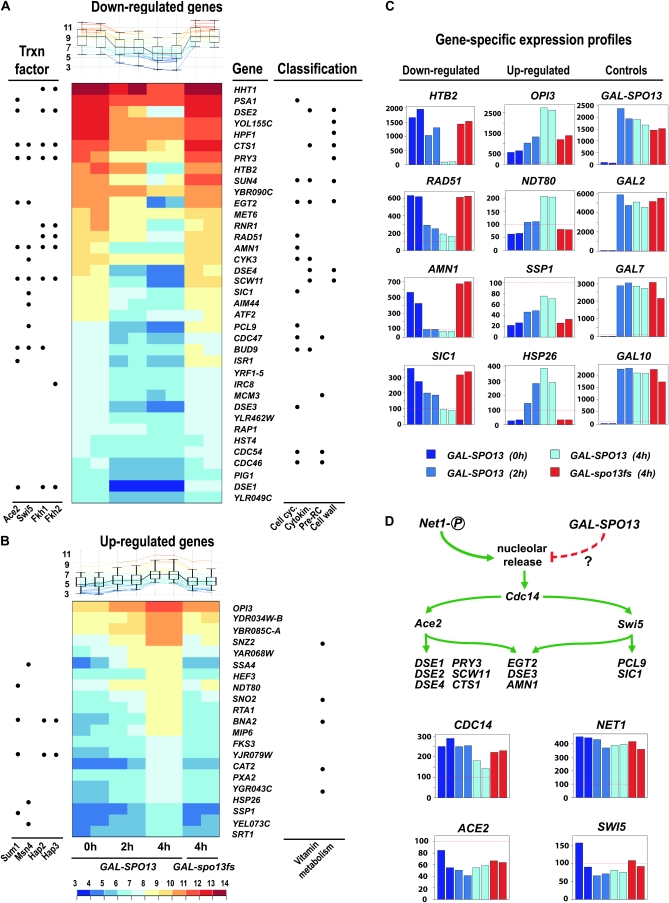
Using a statistical test (ANOVA) with stringent parameters, we identified 58 genes that displayed signal changes during the time course (see materials and methods). K-means clustering revealed 37 genes with reduced mRNA concentration and 21 displaying increased transcript concentration in GAL-SPO13 vs. GAL-spo13fs cells (Figure 3, A and B). The cluster of genes for which reduced signals were obtained (for convenience referred to as downregulated genes, irrespective of whether direct or indirect effects cause their signal reduction) included loci involved in cell-cycle progression (AMN1, DSE3, PCL9, and SIC1), chromatin assembly (HTB2), cytokinesis (BUD9, CTS1, and DSE2), DNA replication (CDC46/MCM5, CDC47/MCM7, CDC54/MCM4, and MCM3), and formation of the cell wall (SCW11) (Figure 3, A and C). The group of genes for which increased mRNA concentrations were detected (referred to as upregulated, regardless of cause or mechanism) included, as expected, SPO13 itself and galactose inducible loci (GAL2, GAL7, and GAL10) (Figure 3C). In addition, we detected genes encoding metabolic functions (BNA2, CAT2, OPI3, SNO2, and SNZ2), a heat-shock protein (HSP26), and NDT80, which encodes a sporulation-specific transcription factor required for middle-meiotic gene expression and activation of its own expression via a positive feedback loop (Pak and Segall 2002). In this context, it is noteworthy that three of the upregulated genes (NDT80, BNA2, and YJR079W) were previously reported to be repressed by Sum1, a DNA-binding transcription factor that represses Ndt80 and a subset of Ndt80-induced genes during vegetative growth (Hepworth et al. 1998; Xie et al. 1999; McCord et al. 2003; Jolly et al. 2005). However, the SUM1 transcript itself is not altered under these conditions, suggesting that the protein may be regulated by post-transcriptional mechanisms.
Figure 3.—
Genome-wide mRNA profiling during Spo13-dependent mitotic arrest. (A and B) Heat maps and graphical displays with signal distributions for each time point, displayed in duplicate. Data are shown for overexpression of wild type (GAL-SPO13, GA-3419) at 0 hr, 2 hr, and 4 hr, and frameshift (GAL-spo13fs, GA-4318) at 4 hr. Left dots indicate genes whose expression depends upon the specific transcription factor given at bottom. Right dots indicate participation in the biological process (given at bottom). Log2-transformed signal intensities are color coded as given in the scale. (C) Bar diagrams summarize linear signal intensities (y-axis) vs. color-coded samples (x-axis) for typical cases of down- and upregulated genes, as well as control genes. The samples are described below the graphs. (D) Model of the pathways that contribute to the Spo13-dependent sequestration or Net1-dependent release of Cdc14 from the nucleolus. Diagram shows regulation of Ace2 and Swi5 transcription factors by Cdc14, which leads to the transcription of their target genes (top). Bar diagrams below show linear expression signals for four selected genes; samples are color coded as in C.
A substantial number of genes whose mRNA concentration dropped below the threshold of detection during Spo13-induced cell-cycle arrest contain binding sites for the DNA-binding transcription factors Ace2 combined with Swi5, Fkh1, or Fkh2 (AMN1, DSE2, and SCW11), or Swi5 alone (PCL9 and SIC1). Since the mRNAs of these genes are not normally downregulated below detection limits of microarrays at the G2/M phase of the mitotic cell cycle, the decreasing signal intensitites are likely due to impaired activator function and not simply a consequence of cells accumulating at the arrest stage (Cho et al. 1998; Granovskaia et al. 2010). We suspected these transcription factors might be post-translationally regulated since neither ACE2 nor SWI5 transcript levels were detectably altered (Figure 3D). Indeed, the nuclear localization of Ace2 and Swi5 is known to be promoted by Cdc14 phosphatase and Net1 phosphorylation during the cell cycle (Visintin et al. 1998; O'Conallain et al. 1999). These results, coupled with the Spo13-induced cell-cycle arrest phenotype shown in Figure 1 (see also McCarroll and Esposito 1994), led us to propose that Spo13 may delay mitotic exit by antagonizing the release of Cdc14 from its nucleolar inhibitor Net1, thereby preventing the nuclear localization of Ace2 and Swi5. This explanation would not only account for the decreased mRNA concentration of their target genes (Figure 3D), but would provide a mechanism for the Spo13-induced mitotic arrest, given the important role of Cdc14 phosphatase in progression through mitosis.
Spo13 overexpression inhibits nucleolar release of Cdc14 phosphatase:
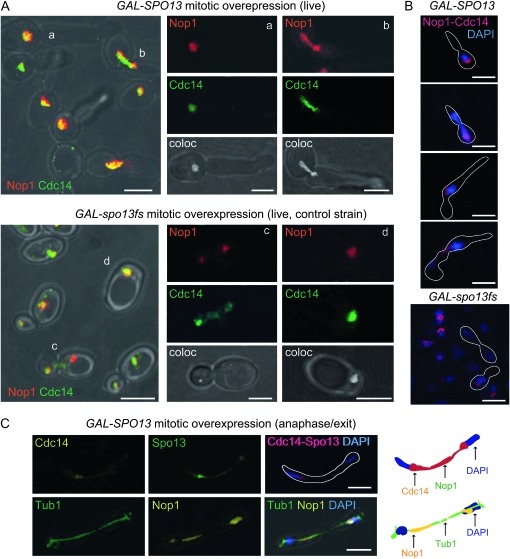
To test the status of Cdc14 activation upon Spo13 overexpression, we introduced a CDC14-GFP gene fusion into GAL-SPO13 or GAL-spo13fs cells for live analysis. The nucleolus was visualized by expression of a plasmid-borne NOP1-CFP fusion (Bystricky et al. 2005). After a 2-hr induction of either GAL-SPO13 or GAL-spo13fs, cells were examined by confocal microscopy. In Spo13-arrested cells, Cdc14 colocalized with Nop1 in the mother cell nucleolus (Figure 4A, a; G2/M arrest phenotype), or in the rDNA domain spanning from mother to daughter nuclei in anaphase (Figure 4A, b). There was no detectable release of Cdc14. In contrast, in cells expressing spo13fs, Cdc14 was released from its interphase colocalization with the nucleolus (Nop1) in late mitosis, and cells continued through the cell cycle (Figure 4A, c and d).
We confirmed the live-imaging phenotypes by anti-HA immunostaining of HA-tagged Cdc14 in strains expressing either GAL-SPO13 or GAL-spo13fs. In all cells blocked by the Spo13-induced arrest, we found Cdc14-HA colocalized with Nop1 (Figure 4B). Strikingly, even in transition and A/exit cells in which the nucleolus spans mother and daughter cells, Cdc14 remained nucleolar. Again, a significant number of these cells (19% of total cells scored; n = 130) had anaphase spindles (Figure 4C, anti-tubulin), providing further evidence for the second point of arrest or delay in late anaphase in response to high Spo13 levels (Table 2). Importantly, in all cases Spo13-expressing cells arrested with Cdc14 in the nucleolus, unlike late anaphase cells expressing spo13fs (Figure 4). These data support the conclusion that Spo13 directly or indirectly interferes with Cdc14 release and thereby delays the downregulation of the mitotic Cdk, an event essential for the A/exit transition.
TABLE 2.
Spo13 inhibits Cdc14 release from the nucleolus
| Cdc14-Nop1 colocalization/number examined | |||||
|---|---|---|---|---|---|
| Strain | Exp | G1 | S | G2/M | Extended nucleolus |
| GAL-spo13fs | Live | 184/184 | 86/86 | 7/7 | 0/5 |
| GAL-SPO13 | Live | — | — | 66/66 | 12/12 |
| GAL-spo13fs | IF | 43/43 | 45/45 | 22/22 | 0/22 |
| GAL-SPO13 | IF | — | — | 93/93 | 17/20 |
A summary of the frequency of observed phenotypes after 2 hr of induction of GAL-SPO13 on galactose from the experiments shown in Figure 4, A and B. Experimental details are described in the legend to Figure 4. IF, immunostaining; live, live GFP imaging.
Spo13 overexpression affects the stability of Clb2:
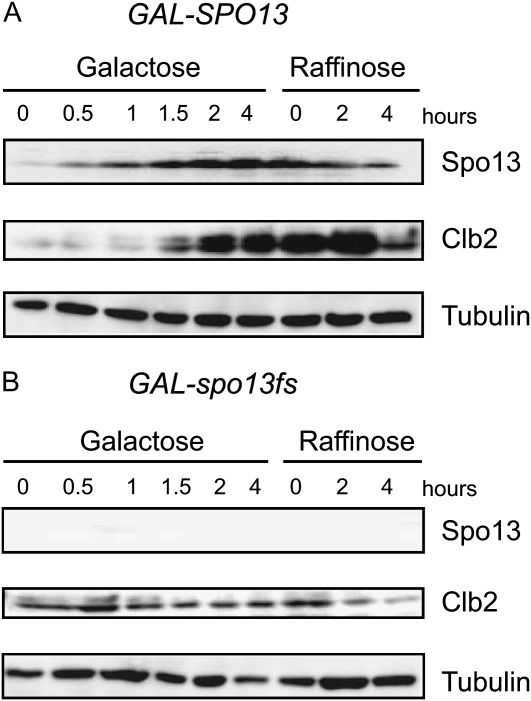
To examine the molecular effects of Spo13 overexpression further, we monitored the reversibility of changes at a biochemical level. After 4 hr on galactose, strains harboring GAL-SPO13 and GAL-spo13fs were shifted back to glucose and released into a medium containing raffinose, in which the GAL1 promoter remains repressed. Samples were taken at the indicated time points, and the instability of Clb2, which normally increases as cells progress through mitosis, was assayed by Western blot (Figure 5). As expected, the Spo13 protein accumulated during GAL-SPO13 induction and was not present in cells expressing the frameshift allele (spo13fs; Figure 5, A and B). Spo13 persisted, albeit at a reduced level, for 4 hr even after the GAL1 promoter was repressed, consistent with it being stabilized by the M-phase Cdk. Clb2 also accumulated to high levels during Spo13-induced arrest (Figure 5A; see Lee et al. 2002) and remained high during 4 hr on raffinose, suggesting that APC-dependent degradation of Clb2 was impaired. Neither the accumulation of Clb2 on galactose, nor its degradation once Spo13 protein levels decreased, were observed in GAL-spo13fs cells (Figure 5B)
Both Cdc14 activation and a functional APC are necessary for Clb2 degradation in late mitosis (Stegmeier and Amon 2004; Queralt et al. 2006; Toth et al. 2007). Thus the persistent accumulation of Clb2 in the presence of overexpressed Spo13 is consistent with the proposal that a high level of Spo13 in mitosis inhibits Cdc14 activation and directly or indirectly impairs activation of the APC.
ESP1 separase overexpression bypasses the cell-cycle arrest imposed by Spo13:
To determine how Spo13 might inhibit Cdc14 release, we investigated the relationship between overexpressed Spo13 and the separase Esp1. Besides its role in cleaving cohesin, Esp1 functions in the FEAR (Stegmeier et al. 2002) and MEN (Queralt et al. 2006) pathways, leading to the phosphorylation of Net1 and subsequent release of Cdc14 from the nucleolus, which in turn promotes exit from mitosis. Esp1 acts in this context by inactivating PP2ACdc55 phosphatase to allow phosphorylation of both Net1 by Cdc28 kinase (FEAR activation) and the GAP complex (Bub2/Bfa1) promoting MEN activation (Queralt et al. 2006). Intriguingly, Esp1 overexpression is sufficient to trigger Cdc14 activation, through a mechanism independent of its proteolytic activity on cohesin (Sullivan and Uhlmann 2003). To distinguish whether Spo13 interferes with Cdc14 release by acting upstream or downstream of Esp1, we used similar suppression analyses.
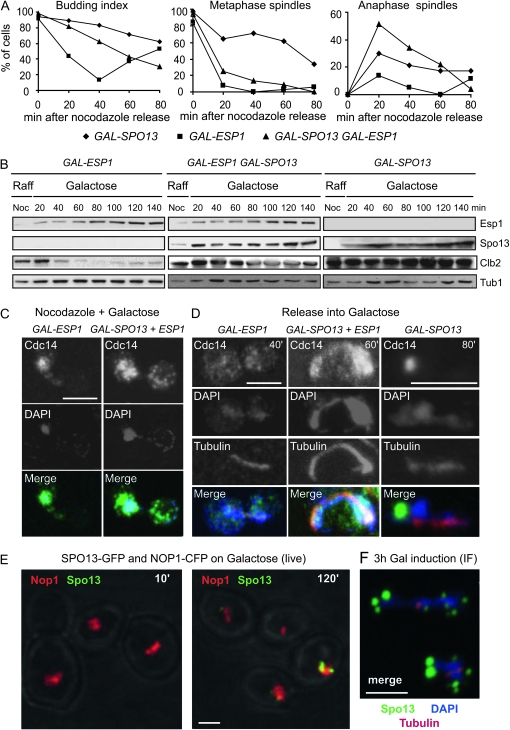
Using strains carrying either GAL-SPO13, GAL-ESP1, or both, we tested whether SPO13 induction overrides the release of Cdc14 provoked by GAL-ESP1 induction (Tinker-Kulberg and Morgan 1999; Sullivan and Uhlmann 2003). To ensure that cells have progressed properly through S phase without premature sister separation, we blocked cells at the G2/M boundary by exposure to nocodazole under noninducing conditions. Galactose was then added for 1 hr, cells were washed with YP raffinose, and released from nocodazole into galactose. Cell-cycle progression was monitored by both fluorescence imaging and Western blot analysis (Figure 6, A–D). As expected, GAL-ESP1 cells passed through mitotic exit and began to rebud 40 min after release, coinciding with the degradation of Clb2. In contrast, GAL-SPO13 cells remained arrested, with high Clb2 and Spo13 levels. The persistence of metaphase and anaphase spindles throughout the experiment in these cells agreed with our earlier results on mitotic arrest.
Figure 6.—
ESP1 overexpression suppresses Spo13-induced arrest. (A) Graphs show cell counts for budding index, metaphase or anaphase spindles on strains expressing GAL-SPO13, GAL-ESP1, and both GAL-SPO13 and GAL-ESP1 as well as CDC14-6HA (GA-5441, GA-5406, and GA-5230, respectively). Cells were blocked at G2/M with nocodazole and released in the presence of galactose. (B) Western blot analysis of the indicated proteins at the indicated time points from the experiment described in A. (C and D) Representative micrographs showing immunofluorescence analysis of the indicated proteins and strains either during galactose induction on nocodazole (C) or on cells released from nocodazole arrest for 40, 60, or 80 min on galactose (D). GAL-SPO13 refers to strain GA-5441, GAL-ESP1 is GA-5406, and GAL-SPO13 + ESP1 is GA-5230. Quantitation of Cdc14 release at equivalent time points is as follows: Under nocodazole arrest with overexpression of ESP1, 83%; SPO13, 19%; ESP1 SPO13, 55%; after arrest and release into galactose, ESP1, 97% (at 40 min); SPO13, 20% (at 80 min); and ESP1 SPO13, 56% (at 60 min). Between 30 and 106 cells were counted for each point. (E) Representative live fluorescence images of strain GAL-SPO13-GFP pNOP1-CFP (GA-4175) showing Nop1-CFP (red) and Spo13-GFP (green) at 10 and 120 min after shift to galactose-containing media. Bar, 2 μm. A total of 85% of cells counted show colocalization. (F) After 3 hr induction on galactose some mitotic cells show large aggregates of Spo13-GFP that do not colocalize with DNA, the nucleolus nor the spindle, detected here by immunofluorescence (red, tubulin; green, Spo13; blue, DAPI). Bar, 2 μm.
In contrast to overexpression of Spo13 alone, the coordinated overexpression of both Esp1 and Spo13 allowed metaphase spindles to disappear quickly despite the presence of high levels of Spo13. Thus, Esp1 overexpression clearly suppressed the Spo13-induced arrest at G2/M. These cells also accumulated anaphase spindles rapidly after release from nocodazole, illustrating the effect of Spo13 on anaphase/exit, which was less visible in asynchronous cells. By 80 min the anaphase spindles were disassembled, demonstrating more rapid progression in the presence of both Esp1 and Spo13, compared to the presence of Spo13 alone (Figure 6A). Consistently, Clb2 degradation also occurred faster in strains overexpressing both genes than in cells with GAL-SPO13 only (Figure 6B). These data indicate a suppression of the Spo13-induced arrest by Esp1, which is known to confer Cdc14 release and activation. Interestingly, in the ESP1 SPO13 double-expressing strain low levels of Clb2 were still detectable at 80, 100, and 120 min, consistent with incomplete APC activation.
Immunofluorescence studies performed for Cdc14 on these same cells showed that Cdc14 is released both in nocodazole-arrested and in anaphase cells in the presence of Esp1, either when expressed alone or together with Spo13 (Figure 6, C and D). As expected, cells expressing GAL-SPO13 alone accumulated anaphase spindles, and Cdc14 release from the nucleolus was less efficient (Figure 6D). On galactose, Cdc14 was released in 97% of cells when Esp1 was overexpressed alone, in 20% of cells when Spo13 was overexpressed alone, and in 56% of cells when both were expressed. Given that the expression of Esp1 overrides the Spo13-induced arrest at both stages of mitosis, and reverses the Spo13-induced sequestration of Cdc14, these data strongly suggest that Spo13 acts upstream of Esp1 (or in parallel to it) to regulate both the FEAR (G2/M) and MEN (A/exit) pathways.
Finally, to test whether Spo13 localization provides any further insight into its mode of action, we monitored the subcellular distribution of overexpressed Spo13 in living cells. At 2 hr of induction of GAL-SPO13-GFP ∼80% of the Spo13-GFP foci colocalized with the nucleolar marker Nop1 (n = 120; Figure 6E, right), providing novel evidence for its association with the nucleolus. The distribution of the Spo13 signal was nonetheless distinct from the uniform nucleolar staining of Cdc14 (Figure 6E). By 3 hr of induction multiple large nuclear foci of Spo13-GFP were observed, which did not colocalize with either the mitotic spindle or the nucleolus (Figure 6F). The significance of this localization with respect to Esp1 and Cdc14 activation remains unclear.
Spo13 interferes with the APC pathway:
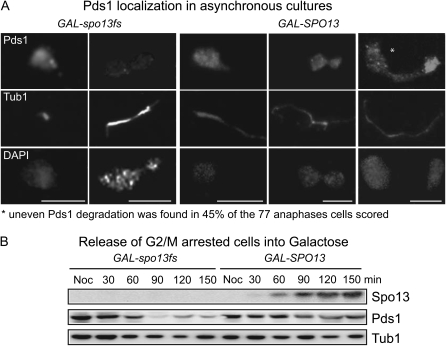
The above data suggest that Spo13 may act upstream of separase (Esp1) to impair the release of Cdc14 from its inhibitor Net1, which is required for progression through mitosis. Given that separase is activated by APC cleavage of securin (Pds1) (Cohen-Fix et al. 1996), it seems a plausible hypothesis that APC function is diminished in the presence of Spo13 (Katis et al. 2004). To test this, we investigated the stability of securin (Pds1), a key APC substrate, during Spo13 overexpression. Pds1 was tagged with 18-Myc epitopes in strains carrying GAL-SPO13 or GAL-spo13fs. After 4 hr of galactose induction, samples were taken and processed for immunofluorescence (Figure 7A). In the spo13-fs strain, Pds1 was present in G2/M cells but not detectable in cells at anaphase. In contrast, Pds1 was detectable at all cell-cycle phases in cells overexpressing Spo13. Intriguingly, cells with long spindles usually showed a heterogeneous pattern, with Pds1 signal being stronger in one of the two stretched nuclei. This suggests that Pds1 is partially degraded over time and/or is being resynthesized in a nucleus that has progressed further in the exit pathway.
We examined this further by studying Pds1 stability in synchronized cultures. The GAL-SPO13 and GAL-spo13-fs strains were grown in YP+raffinose to reach mid-log phase (5 × 106 cells/ml), at which point, nocodazole was added for 1.5 hr to block cells in G2/M. Cells were then washed twice with YP-raffinose, released into drug-free YP-galactose to induce Spo13 expression, and samples analyzed every 30 min and analyzed by Western blot (Figure 7B). In the control strain expressing GAL-spo13fs, Pds1 levels started to drop 60 min after release from nocodazole and were undetectable by 90 min. In contrast, significant amounts of stabilized Pds1 (30–50% of the initial level) were seen in cells expressing Spo13. These results confirm that normal degradation of both Pds1 and Clb2, two key substrates for APC, is inhibited by Spo13 in mitotic cells, providing further support for the notion that Spo13 either directly or indirectly inhibits APC function (Katis et al. 2004).
DISCUSSION
It has been proposed that Spo13 acts as a transient negative regulator or timer, slowing meiotic events to (1) allow proper establishment of the reductional machinery for centromere cohesion/coorientation prior to the first M phase, allowing homologs to segregate properly, and (2) impede the anaphase to MI exit, thereby ensuring two successive meiotic divisions without an intervening S phase. This permits haploid products to form (McCarroll and Esposito 1994). The present study exploited the mitotic overexpression phenotype of Spo13 to probe Spo13's mechanism of action in regulating cell cycle progression.
When expressed in mitotically dividing cells, the Spo13 protein was stabilized in both S- and M-phase cells, and induced a cell-cycle arrest that is largely independent of the spindle checkpoint gene MAD2. Importantly, Spo13 does not accumulate to detectable levels in G1-arrested cells, and Spo13-induced arrest can be reversed by repressing Spo13 expression or provoking Spo13 degradation through inhibition of the Cdc28/Clb kinase complex. The use of an analog-sensitive Cdc28 gene suggests that Spo13 stability is directly dependent on the mitotic activity of the Cdk/Clb complex. Indeed, Spo13, which has 10 SP/TP consenses, is phosphorylated (Figure 2). This would argue for a reciprocal positive feedback loop between Spo13 and Cdc28. Namely, SPO13 overexpression results in the accumulation of Clb2 and an enhancement of Cdc28/Clb activity, which in turn stabilize Spo13 (McCarroll and Esposito 1994). Consistent with this model, a recent report suggests that Spo13 is a target of APC, which may degrade unphosphorylated Spo13 in G1 (Sullivan and Morgan 2007).
Our data provide compelling evidence that Spo13 delays mitotic cell-cycle progression at two different points: at the G2/M transition and late in anaphase. The effect at anaphase is particularly clear if Spo13 is induced in synchronized cells released from a metaphase arrest. We see enrichment of anaphase spindles and poor degradation of Pds1 securin by both immunofluorescence and Western blot analysis. These findings suggest that the level of Pds1 degradation that occurs in the presence of Spo13 is insufficient to allow the Esp1-mediated cohesin cleavage needed for the G2/M transition, as well as downregulation of PP2ACdc55, which is necessary for Cdc14 release from the nucleolus and the segregation of rDNA at anaphase (Sullivan et al. 2004).
Genome-wide microarray analysis provided useful clues as to how Spo13 overexpression might arrest cells in late anaphase. We observed progressively decreasing mRNA concentrations for a number of cell cycle-regulated genes that are under the control of Ace2 and Swi5 transcription factors, including some cases where the mRNAs became undetectable in arrested cells. This is not seen in other G2/M-arrested cells. Both Ace2 and Swi5, which are substrates of the Cdc14 phosphatase, require dephosphorylation during late anaphase to activate their function (F. Uhlmann, personal communication). This led us to examine whether Spo13 might inhibit A/exit by negatively regulating Cdc14. Indeed, we find that ectopic expression of Spo13 prevents the release of Cdc14 from the nucleolus, an event that correlates with Cdc14 activation and subsequently with exit from mitosis (Stegmeier et al. 2002; Buonomo et al. 2003; Marston et al. 2003). Given that the proper balance of Cdc14 and Cdk/Clb activities is known to be crucial for triggering mitotic exit (Toth et al. 2007), we conclude that high levels of Spo13 must also ultimately interfere with this balance.
Of special relevance for interpreting the effects of Spo13 is the finding that separase (Esp1) plays a key role in coordinating cohesin cleavage at G2/M with subsequent Cdc14 release during anaphase. Esp1 not only cleaves Scc1 releasing cohesion, but also downregulates PP2ACdc55, thus allowing Net1 phosphorylation (Queralt et al. 2006) and Cdc14 release (Shou et al. 1999; Visintin et al. 1999). On the basis of this finding, we tested whether the effects of Spo13 overexpression might act in part by reducing Esp1 function. Indeed, ESP1 overexpression can bypass the Spo13-induced G2/M arrest as well as the nucleolar retention of Cdc14, arguing that Spo13 either acts upstream or in parallel to Esp1 function. Since ESP1 overexpression also overcomes metaphase arrest in the absence of APC (Uhlmann et al. 2000), our data suggest that Spo13 may modulate Esp1 activity by inhibiting APC (see also Lee et al. 2002; Katis et al. 2004). We tested this by monitoring the degradation of a key APC target, securin, or Pds1. We could confirm that high levels of Spo13 impair Pds1 degradation. This in turn would prevent complete activation of separase (Esp1), which is needed for both anaphase progression and exit from mitosis.
It is important to note that while Esp1 overexpression could overcome Spo13 inhibition to allow Cdc14 release, it did not fully rescue the delay in the A/exit transition resulting from Spo13 overexpression. During the combined overexpression of both ESP1 and SPO13, Clb2 persisted in a stable state longer than in the strain overexpressing ESP1 alone (Figure 6B). This suggests that Spo13 has additional effects on A/exit, independent of separase. One plausible explanation is that Spo13 may directly or indirectly inhibit APC function, which acts both upstream and downstream of separase. It has recently become clear that Cdc14 activation promotes exit from mitosis by allowing dephosphorylation of the APC regulator Cdh1 as well as a set of late-anaphase substrates (Visintin et al. 1998; Stegmeier et al. 2002; D'amours et al. 2004; Stegmeier and Amon 2004; Sullivan et al. 2004; Torres-Rosell et al. 2005; Queralt et al. 2006; Khmelinskii et al. 2007; Toth et al. 2007). The failure to release Cdc14 in cells overexpressing Spo13 and the massive accumulation of Clb2 in late anaphase in these cells suggests that Spo13 also negatively regulates anaphase by altering the late mitotic balance between Cdc14 and Cdc28/Clb2 activities as indicated earlier.
How does this mechanism relate to Spo13's role in meiotic division? As noted earlier, in meiosis Spo13 is required for homolog separation and two successive M phases, MI and MII, without an intervening S phase. The meiotic Spo13 protein is detected specifically during MI when its transcript level peaks, and it is degraded by APC at the end of MI. Loss of SPO13 function causes diploid cells (or haploids expressing both mating type alleles), to undergo a single, largely equational, division on the MI spindle and exit the cell cycle forming dyads with two diploid (or haploid) spores (Klapholz and Esposito 1980b; Wagstaff et al. 1982; Rutkowski and Esposito 2000). The termination of meiosis after a single division, in which many chromosomes separate sister chromatids instead of homologs, appears to depend on the Mad2 spindle checkpoint (Shonn et al. 2000). The question remains, however, why wild-type Spo13 cells, which undergo a reductional MI, execute a second M phase without first exiting division and initiating another round of DNA replication. We propose that Spo13 may be able to promote MII immediately after MI, by preventing the complete loss of Clb/Cdk activity and the A/exit transition (Queralt et al. 2006; Toth et al. 2007). Only after mitotic exit can prereplication complexes assemble to allow a subsequent round of DNA replication. The ability of Spo13 to prevent a second S phase by keeping Clb/Cdk activity high is consistent with the fact that overexpression of SPO13 in the cdc28-1 mutant at a semi-permissive temperature (30°) allowed MII to immediately follow MI in meiotic cells as described earlier (Shuster and Byers 1989; McCarroll and Esposito 1994). In this case, we speculate that Spo13 modulates Cdk activity by maintaining high cyclin B levels (Figure 6) and preventing full Cdc14 release from the nucleolus. As discussed above, this could be achieved by interfering with APC function, although proof of this requires future analysis. Consistent with this model, we note that yeast cyclins have been detected during meiosis throughout MI and MII (Carlile and Amon 2008).
The control of two successive chromosome segregation events (MI and MII) without an intervening S phase is a fundamental feature of meiosis, which is conserved from yeast to man. The fact that in meiotically dividing frog oocytes cyclin B appears not to be degraded between MI and MII (Taieb et al. 2001) provides a tantalizing hint that the effects of Spo13 on anaphase and exit from mitosis in yeast and could well provide a plausible model for how organisms universally control this crucial event.
Acknowledgments
We thank P. Demougin and M. Cartron for help with yeast array experiments, M. Rebhan for protein motif analysis and F. Uhlmann for kindly providing strains that were used to generate those used here. Finally, R.E.E. extends special thanks to S.M.G. and M.P. for graciously hosting her during several sabbatical stays in Basel. E.V. was supported by the Swiss Cancer League, the Novartis Research Foundation, and a short-term European Molecular Biology Organization fellowship. This work was further funded by National Institutes of Health GM-29182 (to R.E.E.) and Swiss National Science Foundation grant 3100A0-105861 (to M.P.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.113746/DC1.
References
- Azzam, R., S. L. Chen, W. Shou, G. Alexandru, K. Nasmyth et al., 2004. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305 516–519. [DOI] [PubMed] [Google Scholar]
- Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray et al., 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407 395–401. [DOI] [PubMed] [Google Scholar]
- Brown, N. R., M. E. Noble, J. A. Endicott and L. N. Johnson, 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1 438–443. [DOI] [PubMed] [Google Scholar]
- Buckingham, L. E., H. T. Wang, R. T. Elder, R. M. McCarroll, M. R. Slater et al., 1990. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87 9406–9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo, S. B., K. P. Rabitsch, J. Fuchs, S. Gruber, M. Sullivan et al., 2003. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev. Cell 4 727–739. [DOI] [PubMed] [Google Scholar]
- Bystricky, K., T. Laroche, G. van Houwe, M. Blaszczyk and S. M. Gasser, 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile, T. M., and A. Amon, 2008. Meiosis I is established through division-specific translational control of a cyclin. Cell 133 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel, F., and M. Primig, 2008. The Annotation, Mapping, Expression and Network (AMEN) suite of tools for molecular systems biology. BMC Bioinformatics 9 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway et al., 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2 65–73. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O., J. M. Peters, M. W Kirschner and D. Koshland, 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10 3081–3093. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., F. Stegmeier and A. Amon, 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117 455–469. [DOI] [PubMed] [Google Scholar]
- Gattiker, A., C. Niederhauser-Wiederkehr, J. Moore, L. Hermida and M. Primig, 2007. The GermOnline cross-species systems browser provides comprehensive information on genes and gene products relevant for sexual reproduction. Nucleic Acids Res. 35 D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat, M., S. Jensen and L. H. Johnston, 2002. Mitotic exit: the Cdc14 double cross. Curr. Biol. 12 R482–R484. [DOI] [PubMed] [Google Scholar]
- Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan et al., 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granovskaia, M. V., L. J. Jensen, M. E. Ritchie, J. Toedling, Y. Ning et al., 2010. High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol. 11 R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth, S. R., H. Friesen and J. Segall, 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18 5750–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen, A., G. Wrobel, M. Cartron, P. Demougin, C. Niederhauser-Wiederkehr et al., 2005. Novel response to microtubule perturbation in meiosis. Mol. Cell. Biol. 25 4767–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly, E. R., C. S. Chin, I. Herskowitz and H. Li, 2005. Genome-wide identification of the regulatory targets of a transcription factor using biochemical characterization and computational genomic analysis. BMC Bioinformatics 6 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis, V. L., J. Matos, S. Mori, K. Shirahige, W. Zachariae et al., 2004. Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr. Biol. 14 2183–2196. [DOI] [PubMed] [Google Scholar]
- Khmelinskii, A., C. Lawrence, J. Roostalu and E. Schiebel, 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz, S., and R. E. Esposito, 1980. a Isolation of SPO12–1 and SPO13–1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 96 567–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz, S., and R. E. Esposito, 1980. b Recombination and chromosome segregation during the single division meiosis in SPO12–1 and SPO13–1 diploids. Genetics 96 589–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S. B. Buonomo, C. Michaelis et al., 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 91–103. [DOI] [PubMed] [Google Scholar]
- Lee, B. H., A. Amon and S. Prinz, 2002. Spo13 regulates cohesin cleavage. Genes Dev. 16 1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B. H., B. M. Kiburz and A. Amon, 2004. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr. Biol. 14 2168–2182. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Marston, A. L., B. H. Lee and A. Amon, 2003. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell 4 711–726. [DOI] [PubMed] [Google Scholar]
- McCarroll, R. M., and R. E. Esposito, 1994. SPO13 negatively regulates the progression of mitotic and meiotic nuclear division in Saccharomyces cerevisiae. Genetics 138 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord, R., M. Pierce, J. Xie, S. Wonkatal, C. Mickel et al., 2003. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23 2009–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Conallain, C., M. T. Doolin, C. Taggart, F. Thornton and G. Butler, 1999. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak, J., and J. Segall, 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22 6417–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queralt, E., C. Lehane, B. Novak and F. Uhlmann, 2006. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125 719–732. [DOI] [PubMed] [Google Scholar]
- Rabitsch, K. P., M. Petronczki, J. P. Javerzat, S. Genier, B. Chwalla et al., 2003. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell 4 535–548. [DOI] [PubMed] [Google Scholar]
- Rutkowski, L. H., and R. E. Esposito, 2000. Recombination can partially substitute for SPO13 in regulating meiosis I in budding yeast. Genetics 155 1607–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht, U., I. Erb, P. Demougin, N. Robine, V. Borde et al., 2008. Genome-wide expression profiling, in vivo DNA binding analysis and probabilistic motif prediction reveal novel Abf1 target genes during fermentation, respiration and sporulation in yeast. Mol. Biol. Cell 19 2193–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn, M. A., R. McCarroll and A. W. Murray, 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289 300–303. [DOI] [PubMed] [Google Scholar]
- Shonn, M. A., R. McCarroll and A. W. Murray, 2002. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 16 1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed et al., 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 233–244. [DOI] [PubMed] [Google Scholar]
- Shuster, E. O., and B. Byers, 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics 123 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier, F., and A. Amon, 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38 203–232. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., R. Visintin and A. Amon, 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108 207–220. [DOI] [PubMed] [Google Scholar]
- Sullivan, M., and F. Uhlmann, 2003. A non-proteolytic function of the separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 5 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M., T. Higuchi, V. L. Katis and F. Uhlmann, 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117 471–482. [DOI] [PubMed] [Google Scholar]
- Sullivan, M., and D. O. Morgan, 2007. A novel destruction sequence targets the meiotic regulator Spo13 for anaphase-promoting complex-dependent degradation in anaphase I. J. Biol. Chem. 282 19710–19715. [DOI] [PubMed] [Google Scholar]
- Surosky, R. T., and R. E. Esposito, 1992. Early meiotic transcripts are highly unstable in Saccharomyces cerevisiae. Mol. Cell. Biol. 12 3948–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb, F. E., S. D. Gross, A. L. Lewellyn and J. L. Maller, 2001. Activation of the anaphase-promoting complex and degradation of cyclin B is not required for progression from meiosis I to II in Xenopus oocytes. Curr. Biol. 11 508–513. [DOI] [PubMed] [Google Scholar]
- Tinker-Kulberg, R. L., and D. O. Morgan, 1999. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 13 1936–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell, J., F. Machin and L. Aragon, 2005. Cdc14 and the temporal coordination between mitotic exit and chromosome segregation. Cell Cycle 4 109–112. [DOI] [PubMed] [Google Scholar]
- Toth, A., K. P. Rabitsch, M. Galova, A. Schleiffer, S. B. Buonomo et al., 2000. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103 1155–1168. [DOI] [PubMed] [Google Scholar]
- Toth, A., E. Queralt, F. Uhlmann and B. Novak, 2007. Mitotic exit in two dimensions. J. Theor. Biol. 248 560–573. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., D. Wernic, M. A. Poupart, E. V. Koonin and K. Nasmyth, 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103 375–386. [DOI] [PubMed] [Google Scholar]
- Varela, E., K. Shimada, T. Laroche, D. Leroy and S. M. Gasser, 2009. Lte1, Cdc14 and MEN-controlled Cdk inactivation in yeast coordinate rDNA decompaction with late telophase progression. EMBO J. 28 1562–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers et al., 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin, R., E. S. Hwang and A. Amon, 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398 818–823. [DOI] [PubMed] [Google Scholar]
- Wagstaff, J. E., S. Klapholz and R. E. Esposito, 1982. Meiosis in haploid yeast. Proc. Natl. Acad. Sci USA 79 2986–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. T., S. Frackman, J. Kowalisyn, R. E. Esposito and R. Elder, 1987. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol. Cell. Biol. 7 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. M., M. Primig, B. K. Washburn, E. A. Winzeler, M. Bellis et al., 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99 13431–13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter et al., 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]