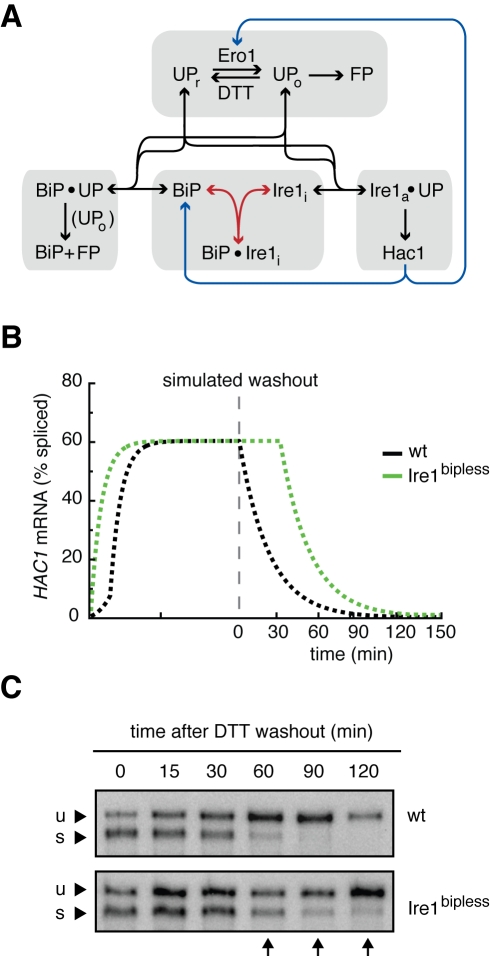
Figure 4. Model architecture, prediction and experimental validation.
(A) The molecular interactions that comprise the model. See the supplement for complete modeling details. Ire1 can exist in three states: (1) inactive monomer (Ire1i, middle lower box), (2) inactive in complex with BiP (Ire1i•BiP, middle lower box), and (3) active in complex with an unfolded protein (Ire1a•UP, lower right box). Either reduced (UPr) or oxidized (UPo) can bind to and activate Ire1, but UPos quickly become folded proteins (FP, upper box and lower left box). The amount of UPrs and UPos is determined by the flux of unfolded proteins and the red/ox potential, defined here as the ratio of Ero1/DTT. Active Ire1 in complex with unfolded proteins produces the Hac1 transcription factor, which induces the production of Ero1 and BiP. BiP can also exist in three states: (1) monomer (BiP, middle lower box), (2) bound to Ire1i (BiP•Ire1i), and (3) in complex with unfolded proteins (BiP•UP). BiP can bind to both UPr and UPo, but only aids in the folding of UPo (bottom left box). The blue arrows indicate the feedback terms that are removed in the “hac1Δ” model, and the red arrows indicate the Ire1/BiP interaction terms that are removed in the “Ire1bipless” model. (B) Simulations “wild type” and “Ire1bipless” cells treated with 5 mM DTT for 100 min and then the DTT is suddenly removed predict a deactivation delay for Ire1bipless cells: “wild type” cells immediately began to deactivate while Ire1bipless continued activity for ∼30 min after DTT withdrawal. (C) Wild type and Ire1bipless were treated with 5 mM DTT for 1 h, filtered, washed, and resuspended in fresh media lacking DTT and sampled over time. Samples were assayed for HAC1 mRNA splicing by Northern blot to measure Ire1 activity. Consistent with the simulations, wild type cells deactivated after 90 min while Ire1bipless cells deactivated after 180 min.