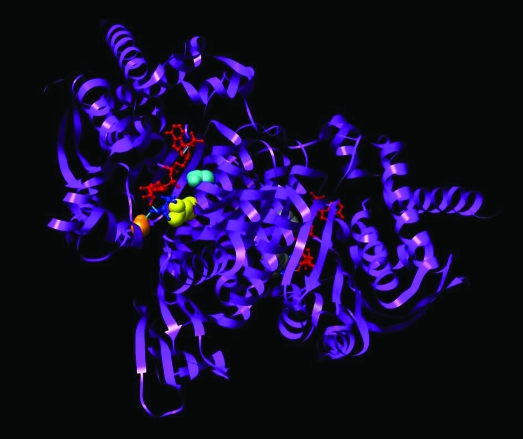
Figure 1.
Isocitrate dehydrogenase 1 (IDH1) dimer in closed conformation. Structure of both molecules of the IDH1 dimer in the active closed conformation. The crystal structure of IDH1 is shown in ribbon format (PDBID:1T0L) (15). The dimer contains two active sites, each of which contains a NADP+-binding site and a metal ion–binding site. One active site is shown in the closed conformation, with the substrate isocitrate in dark blue and the cofactor NADP+ in red. Mutations that alter Arg132 (yellow) to histidine, cysteine, or other amino acids are associated with human gliomas and other cancers. This residue forms three hydrogen bonds with the isocitrate substrate (dark blue). Ser94 (orange) also forms one hydrogen bond with the isocitrate substrate. In the inactive open conformation (data not shown), Asp279 (cyan) contacts Ser94 and sterically hinders isocitrate binding. To transition to the active closed conformation shown here, Asp279 must swing away from Ser94 to relieve this steric hindrance. During this transition, Asp279 contacts Arg132 (15), suggesting that Arg132 plays a role in the transition between inactive and active enzyme conformations. Displayed image was created with University of California San Francisco Chimera software version 1.3 (San Francisco, CA) (16).