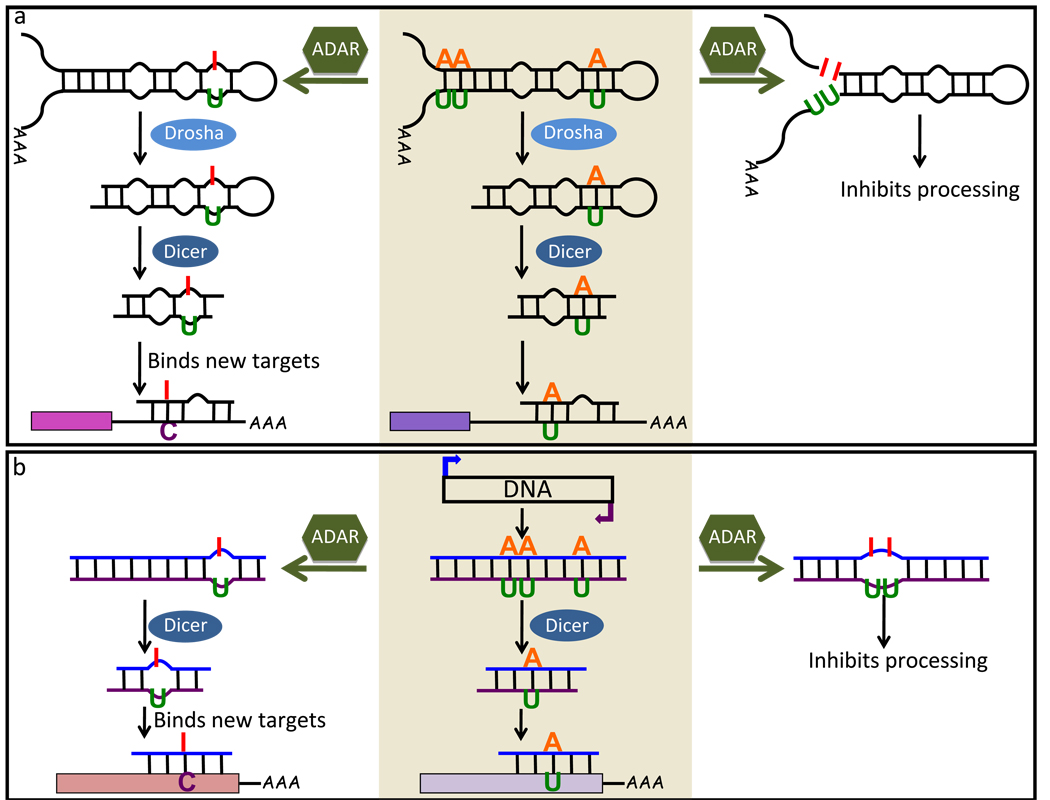
Figure 2.
Models proposed to explain effects of ADARs on dsRNA-mediated gene silencing. miRNA (a) and siRNA (b) pathways are illustrated with precursors, intermediates and mature small RNAs represented as base-paired molecules. Middle pathways (shaded in light brown) show processing by Drosha (light blue oval) and/or Dicer (dark blue oval) in the absence of ADAR (green hexagon), and outer panels show effects of ADARs on processing (right) or re-targeting (left). Representative AU base-pairs are shown being converted to IU mismatches. (a) Drosha processes pri-miRNAs into ~70 nucleotide pre-miRNAs that are then processed into ~22 nucleotide miRNAs by Dicer; as shown, the mature miRNA strand then pairs with the target mRNA [53]. Editing events are detected at a variety of positions in pri-miRNAs, and inosines within both pri- and pre-miRNAs inhibit in vitro cleavage by Drosha and Dicer, respectively [42, 50]. In addition, inosine in at least one endogenous mature miRNA affects binding to mRNA targets [58]. (b) Transgenes that give rise to sense and antisense transcripts produce dsRNA that is edited by ADARs [62]. Inosines within such exogenous dsRNA, or within in vitro transcripts, inhibit processing by Dicer [61]. Although not yet experimentally validated, inosines within siRNAs are predicted to affect binding of siRNAs to target mRNAs. Editing has not been shown to affect the in vivo correlate of this pathway, the endo-siRNA pathway, and there are no reports of endo-siRNAs that contain inosine. However, the characteristics of ADARs make it likely to intersect with the endo-siRNA pathway.