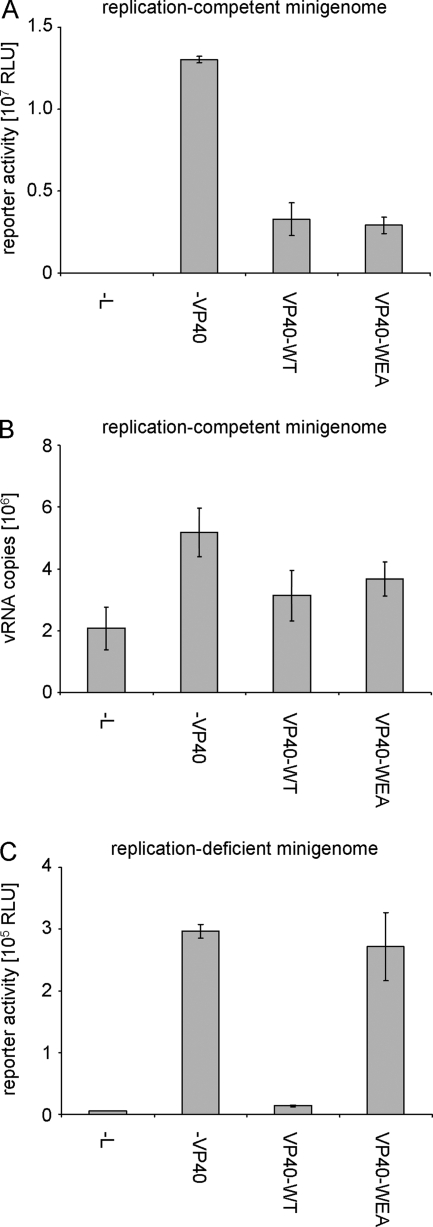
FIG. 6.
Influence of VP40 homooligomerization on viral minigenome replication and transcription. (A) Role in a classical minigenome assay. 293 cells were transfected with a T7-driven minigenome encoding Renilla luciferase, and expression plasmids for the viral proteins NP, L, VP35, VP30, and VP40 (wild type [VP40-WT] or mutant VP40-WEA, as indicated) and accessory plasmids as described in Materials and Methods. Forty-eight hours after transfection, cells were lysed and reporter activity, reflecting both viral replication and transcription, was measured. The average and standard deviation of 3 independent experiments are shown. RLU, relative light units. (B) Influence of VP40 homooligomerization on minigenome replication. 293 cells were transfected with minigenome assay components as described for panel A. Forty-eight hours after transfection, total RNA from these cells was isolated and subjected to a strand-specific quantitative RT-PCR detecting only negative-sense vRNA copies. The average vRNA copy number and standard deviation from 2 independent experiments are shown. (C) Influence of VP40 homooligomerization on transcription of a replication-deficient minigenome. 293 cells were transfected with minigenome assay components as described for panel A; however, instead of a classical minigenome, a replication-deficient minigenome was used. Forty-eight hours after transfection, cells were lysed and reporter activity, reflecting only viral transcription, was measured. The average and standard deviation of 3 independent experiments are shown.