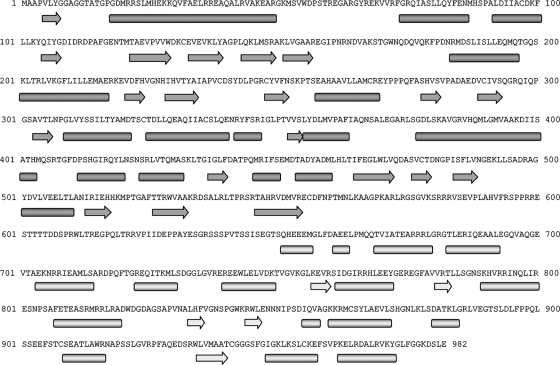
FIG. 4.
Secondary structure consensus prediction for the PcV CP. The PcV CP sequence was obtained from the UniProt database (982 amino acids; accession no. Q8JVC1). Several SSE prediction methods (PsiPred, Jnet, Porter, Sable, Gor, Yaspin, and Profsec) were used to correlate our model of the structural subunit. A consensus SSE prediction was obtained by simple majority at each sequence position. The results were very similar to the consensus SSE prediction obtained with the GeneSilico fold prediction metaserver. The secondary structure gap in the middle of the protein sequence (segment Glu555-Ser650) reflects a structurally disordered region, as confirmed by the consensus prediction of protein order with the GeneSilico metaserver, and divides the PcV coat protein into two parts. Arrow, β-chain; bar, α-helix.