FIG. 8.
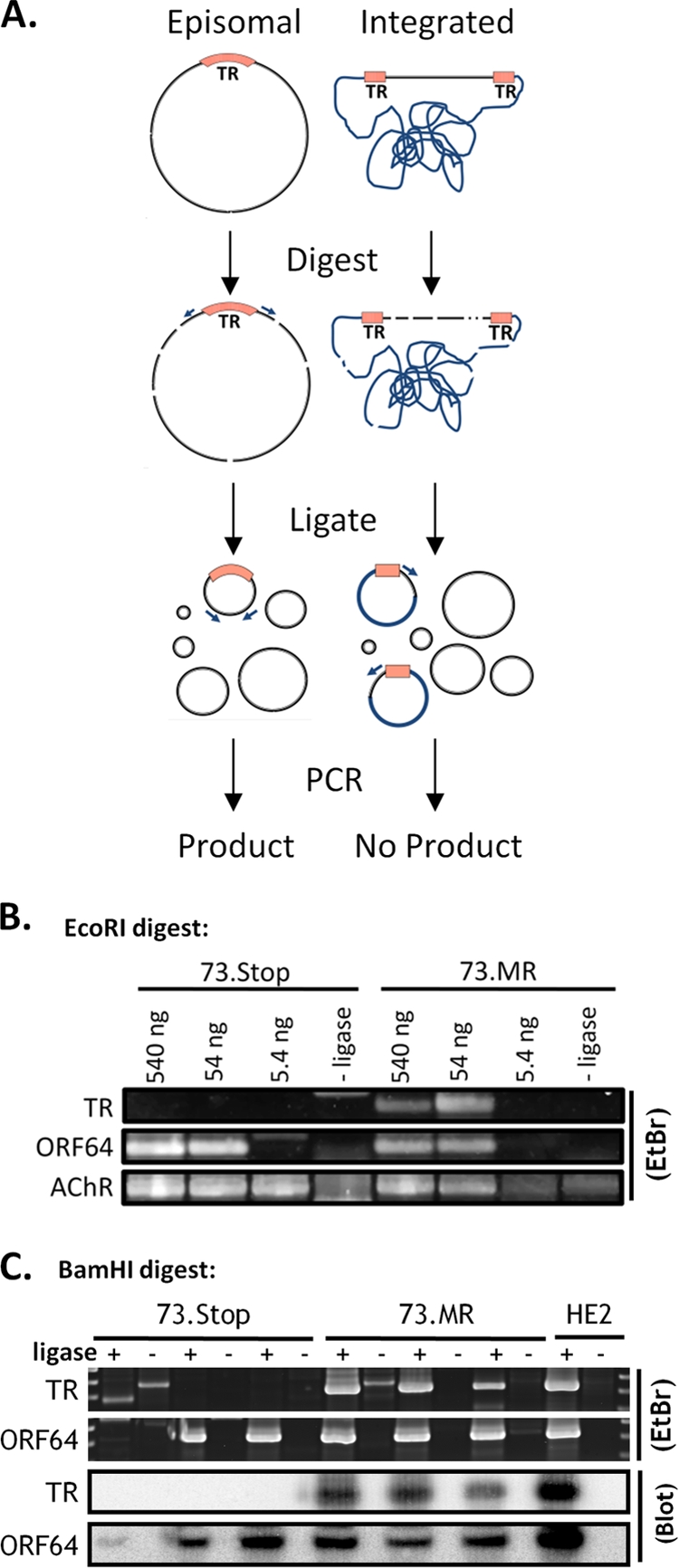
Digestion-circularization PCR reveals mLANA-dependent episome formation during early latency in the spleen. (A) Diagram of the DC-PCR method. Primers are designed facing away from each other on opposite sides of the TR. After digestion and intramolecular circular formation and ligation, the primers will produce a product during PCR. If the virus is integrated, i.e., does not have fused terminal repeats, only one primer binding site will be contained in the resulting circles, and no product is formed. (B) EcoRI digestion and circularization. Numbers above the gel represent the amounts of digested DNA that went into each ligation reaction mixture. One set of mice was analyzed for 73.MR and 73.Stop infections. The murine acetyl choline receptor (AChR) was a control for digestion and intramolecular circularization. (C) BamHI digestion and circularization. Sixty nanograms of DNA was placed into each ligation reaction mixture. Each pair of lanes (ligase +/−) represents a separate group of mice. Ligase-negative reactions are used as a negative control. DNA from the latently infected HE2 (28) cell line was used as a positive control. Southern blots of the PCR products using probes that span the ligated junction further confirmed the specificity of the PCR. EtBr, ethidium bromide.
