Abstract
Lamin A, a key component of the nuclear lamina, is generated from prelamin A by four post-translational processing steps: farnesylation, endoproteolytic release of the last three amino acids of the protein, methylation of the C-terminal farnesylcysteine, and finally, endoproteolytic release of the last 15 amino acids of the protein (including the farnesylcysteine methyl ester). The last cleavage step, mediated by ZMPSTE24, releases mature lamin A. This processing scheme has been conserved through vertebrate evolution and is widely assumed to be crucial for targeting lamin A to the nuclear envelope. However, its physiologic importance has never been tested. To address this issue, we created mice with a “mature lamin A-only” allele (LmnaLAO), which contains a stop codon immediately after the last codon of mature lamin A. Thus, LmnaLAO/LAO mice synthesize mature lamin A directly, bypassing prelamin A synthesis and processing. The levels of mature lamin A in LmnaLAO/LAO mice were indistinguishable from those in “prelamin A-only” mice (LmnaPLAO/PLAO), where all of the lamin A is produced from prelamin A. LmnaLAO/LAO exhibited normal body weights and had no detectable disease phenotypes. A higher frequency of nuclear blebs was observed in LmnaLAO/LAO embryonic fibroblasts; however, the mature lamin A in the tissues of LmnaLAO/LAO mice was positioned normally at the nuclear rim. We conclude that prelamin A processing is dispensable in mice and that direct synthesis of mature lamin A has little if any effect on the targeting of lamin A to the nuclear rim in mouse tissues.
Keywords: Genetic Diseases, Mouse Genetics, Nucleus, Protein Farnesylation, Protein Processing, Gene Knockin, Lamin A, Nuclear Lamina, Prelamin A
Introduction
Lamin A, one of the principal protein components of the nuclear lamina, is generated from prelamin A by a series of four enzymatic post-translational processing steps (1, 2). First, the cysteine in the C-terminal CAAX motif is farnesylated by protein farnesyltransferase. Second, the last three amino acids of the protein (i.e. the -AAX) are clipped off, a redundant activity of two membrane proteases of the endoplasmic reticulum (ER),2 RCE1 and ZMPSTE24 (2, 3). Third, the newly exposed farnesylcysteine is methylated by ICMT (4), a membrane methyltransferase of the ER. Finally, the last 15 amino acids of the protein (including the C-terminal farnesylcysteine methyl ester) are clipped off by ZMPSTE24, releasing mature lamin A. Lamin A and lamin C, both “A-type” lamins, are splice variants of LMNA (5, 6). Lamin C does not contain a CAAX motif and therefore does not undergo any of the C-terminal post-translational processing steps.
The prelamin A processing pathway has attracted considerable attention from medical geneticists, cell biologists, and pharmacologists (1, 7–11). Hutchinson-Gilford progeria syndrome (HGPS), the classic progeroid disorder of children, is caused by point mutations leading to a 50-amino acid internal deletion within the C-terminal region of prelamin A (7, 8). This deletion does not affect protein farnesylation/methylation but abolishes the final cleavage by ZMPSTE24, resulting in the accumulation of a farnesylated, truncated prelamin A in cells (2). This truncated prelamin A, generally called progerin, leads to misshapen nuclei in cells and causes a host of aging-like disease phenotypes (2). A more severe progeroid disorder, restrictive dermopathy (RD), is caused by a deficiency of ZMPSTE24 (12, 13). Without ZMPSTE24, the final cleavage reaction of prelamin A processing cannot occur, preventing mature lamin A synthesis and leading to an accumulation of farnesylated prelamin A (14, 15). The farnesylated prelamin A that accumulates in RD patients is toxic to cells and elicits severe disease (14, 15). Interestingly, several HIV-protease inhibitors block ZMPSTE24 activity, leading to an accumulation of farnesylated prelamin A (9, 10). This prelamin A accumulation conceivably could underlie some side effects of HIV protease inhibitors, for example lipodystrophy and osteoporosis (9, 10).
The discovery of a link between prelamin A processing and progeroid disorders has generated considerable excitement (2, 6, 16, 17). But despite an explosion of interest in prelamin A, some fundamental issues regarding its processing have never been addressed. Notably, it is not even known whether the complicated scheme for lamin A biogenesis is physiologically important. Several cell culture studies have suggested that prelamin A processing is crucial for the delivery of lamin A to the nuclear envelope (18–20), but this issue has never been assessed in vivo. Investigators interested in prelamin A are often asked the following questions: What would happen if the mature form of lamin A were synthesized directly, bypassing all of prelamin A enzymatic modifications? Would the absence of prelamin A processing lead to lethal disease phenotypes, akin to those occurring in Lmna knock-out mice? Is prelamin A processing crucial for the delivery of mature lamin A to the nuclear envelope? The answers to these questions are not known.
We reasoned that prelamin A processing might be essential in mammals because this process has been conserved through vertebrate evolution (19, 21–23), and also because of cell culture studies suggesting that the post-translational processing steps are essential for the targeting of the protein to the nuclear envelope (18–20). We predicted that eliminating prelamin A processing would elicit significant disease phenotypes. To test this prediction, we used gene targeting to create “mature lamin A-only” knock-in mice, where mature lamin A is synthesized directly, bypassing prelamin A processing. We compared the phenotypes of mature lamin A-only mice to “prelamin A-only” knock-in mice, which produce mature lamin A through the normal prelamin A processing pathway. Of note, both mature lamin A-only and prelamin A-only mice lack the capacity to synthesize lamin C. Thus, a side benefit of our experiments was to determine if the synthesis of lamin C is crucial for the growth and vitality of mice.
EXPERIMENTAL PROCEDURES
Generation of Mature Lamin A-only Mice
We generated a mutant Lmna allele yielding only mature lamin A, LmnaLAO, using a strategy similar to one used to generate a mutant Lmna allele yielding progerin (LmnaHG) (32). The arms of the gene-targeting vector were generated by long-range PCR from genomic DNA isolated from 129/OlaHsd embryonic stem (ES) cells. A 3.8-kb 5′ arm, which spanned from the middle of intron 7 to the 3′-untranslated sequences in exon 12, was cloned into pCR2.1-TOPO (Invitrogen). Next, intron 10 was deleted with the QuickChange site-directed mutagenesis kit (Stratagene); that mutation prevents lamin C synthesis. Next, the last 30 nucleotides of exon 11, intron 11, and the first 24 nucleotides of exon 12 were deleted by site-directed mutagenesis; this mutation brings the last codon of mature lamin A immediately adjacent to prelamin A stop codon. The 5′ arm was then cloned into the EcoRI site of pKSloxPNT-mod (33). Next, a 5-kb 3′ arm, identical to the one used in the LmnaHG project (32), was cloned into the AscI site of pKSloxPNT-mod. The vector was linearized with NotI and electroporated into 129/OlaHsd ES cells. Targeted ES cells were identified by Southern blot analysis of EcoRI- or HindIII-digested genomic DNA with a 348-bp 5′-flanking probe. This probe was amplified from mouse genomic DNA with oligonucleotides 5′-CAA GGA GCT CGG ATT CTG TC-3′ and 5′-GTC AGG GAA GAG TGC AGA GG-3′.
Targeted ES cells were microinjected into C57BL/6 blastocysts, and the resulting male chimeric mice were bred with C57BL/6 females to generate heterozygous knock-in mice (LmnaLAO/+). LmnaLAO/+ mice were intercrossed to generate homozygous mice (LmnaLAO/LAO). Genotyping of mice was performed by PCR. The wild-type allele was detected by amplifying a 450-bp fragment with primers 5′-GCT CCC ACT GCC TGT AAC TC-3′ and 5′-CAG AAG ATC ACG TGC AGG A-3′. The mutant allele was detected by amplifying a 350-bp fragment with primers 5′-GCT CCC ACT GCC TGT AAC TC-3′ and 5′-GCC AGA GGC CAC TTG TGT AG-3′.
To exclude the possibility of unwanted mutations in the LmnaLAO or the LmnaPLAO alleles, the full-length lamin A cDNA was isolated from LmnaLAO/LAO fibroblasts, and the full-length prelamin A cDNA was isolated from both LmnaPLAO/PLAO and Lmna+/+ fibroblasts. Aside from the expected mutation in the LmnaLAO transcript, no other mutations were found.
LmnaLAO/+ mice were bred with Zmpste24+/− mice (14, 34) and their offspring intercrossed to obtain Zmpste24+/+Lmna+/+, Zmpste24+/+LmnaLAO/+, Zmpste24−/−Lmna+/+, and Zmpste24−/−LmnaLAO/+ mice. All mice were fed a chow diet and housed in a virus-free barrier facility with a 12-h light-dark cycle. Animal experiments were approved by UCLA Animal Research Committee.
Cell Culture and Western Blot Studies
Primary mouse embryonic fibroblasts were prepared from day 13.5 postcoitum mouse embryos (35). Urea-soluble fibroblast extracts were prepared as described (36), and the proteins were separated on 4–12% gradient polyacrylamide Bis-Tris gels (Invitrogen). The size-separated proteins were transferred to nitrocellulose membranes for Western blotting. The following antibodies were used: goat polyclonal antibodies against lamin A/C (1:400, sc6215), lamin B1 (1:200, sc6217), and actin (1:200, sc1616) (all Santa Cruz Biotechnology), a mouse monoclonal antibody against lamin B2 (1:200, E-3, Invitrogen), and a rat monoclonal antibody against prelamin A (1:50) (37). Secondary antibodies were IRDye 700- or IRDye 800-labeled antibodies against mouse/rat/goat IgG (1:8000, Rockland Immunochemicals). Antibody binding was detected with a Li-Cor Odyssey infrared scanner.
Immunofluorescence Microscopy Studies on Fibroblasts
To assess the morphology of nuclei in embryonic fibroblasts, early passage cells were grown on coverslips, fixed with ice-cold methanol, rinsed briefly with acetone, and stained as described previously (32). The following primary antibodies were used: a rabbit polyclonal antibody against lamin A/C (1:50, sc20681), a goat polyclonal antibody against lamin B1 (1:200, sc6217) (both from Santa Cruz Biotechnology), mouse monoclonal antibodies against lamin A (1:200, MAB3540, Chemicon) and lamin B2 (1:200, E-3, Invitrogen), and a rat monoclonal antibody against prelamin A (1:50) (37). For secondary antibodies, we used Alexa Fluor 488- or Alexa Fluor 568-labeled antibodies against mouse/rabbit/rat/goat IgG (1:200, Invitrogen). DNA was visualized with DAPI. Images were obtained on a Zeiss Axiovert 200M microscope with a ×40 objective and an ApoTome, and processed with AxoVision software, or were obtained on a Leica TCS-SP2 laser scanning confocal microscope with a ×63 objective.
The confocal and STED images (Fig. S3) were recorded with a Leica TCS SP5 STED confocal system equipped with an oil immersion objective (HCX PL APO 100 × 1.4NA STED). A 635-nm pulsed diode laser (PicoQuant, Germany) was used for excitation. The pulses for STED depletion were delivered with a tunable Ti:sapphire laser (Mai Tai BB, Spectra Physics) emitting at 750 nm. Fluorescence emitted from an ATTO 647N-labeled secondary antibody against mouse IgG (Active Motif) was collected through a Semrock BrightLine FF01–685/40–25 nm band pass filter (Rochester, NY) in front of an avalanche photodiode (APD, PerkinElmer). Optical sectioning was achieved by using a detection pinhole set to 1 Airy. Scanning was performed at a line frequency of 10 Hz, averaged 1 to 2 times with an image format of 512 × 512 pixels. Pixel size was kept between 18.5 nm × 18.5 nm and 29.5 nm × 29.5 nm by applying a 10.2–16.4× digital zoom. For comparison of conventional confocal and STED confocal images, all imaging parameters were kept identical except for increasing the APD gain by 7% when recording the STED image in order to compensate for the slight loss of signal in STED microscopy resulting from the shrinking of the effective excitation spot.
In some experiments, primary fibroblasts were treated with an FTI (ABT-100, 2–5 μm) or vehicle (DMSO) alone for 24 h. To analyze the frequency of nuclear blebbing in cultured fibroblasts, >800 cells/genotype were counted in blinded fashion as described (31, 32, 38–40).
Histology and Immunohistochemistry
Paraffin-embedded formalin-fixed tissues from mice were sectioned (5-μm thick) and stained with hematoxylin and eosin. Light microscopy images were taken on a Nikon Eclipse E600 microscope with a ×40 objective and a RT slider 2.3.0 camera; images were recorded with SPOT 4.7 software (Diagnostic Instruments).
To assess the location of lamin A within the nucleus, 6–10 μm-thick frozen tissue sections were fixed with ice-cold methanol, rinsed with acetone, permeabilized with 0.1% Tween-20 in PBS, and then preincubated in phosphate-buffered saline containing 0.1% Tween and 5% fetal bovine serum for 1 h at room temperature. The sections were incubated with a rabbit antibody against lamin A/C (1:50, sc20681, Santa Cruz Biotechnology). After several washes, the sections were incubated for 1 h at room temperature with an Alexa Fluor 568-labeled antibody against rabbit IgG (1:200, Invitrogen). After washes, the sections were fixed with 4% paraformaldehyde and stained with DAPI. Images were obtained with a Zeiss Axiovert 200M microscope equipped with an ApoTome.
Nuclear Strain Experiments
Nuclear strain studies were performed as described previously (25, 29, 31), with the following modifications. Two parallel strips of Scotch tape were attached ∼5 mm apart to the bottom of the silicone membrane of the strain dish, resulting in a uniaxial strain field between the two strips during strain application. Fibroblasts were plated on fibronectin-coated silicone membranes in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS) and cultured for 24 h. Before each assay, cells were incubated with Hoechst 33342 nuclear stain (1.5 μg/ml) for 15 min; the medium was then replaced with phenol red-free DMEM containing 10% FCS and 25 mm Hepes. Membranes were placed on a custom-made strain device, and 20% uniaxial strain was applied to the silicone membrane. Membrane and nuclear strains were computed based on bright field and fluorescent images acquired before, during, and after strain application. Normalized nuclear strain was defined as the ratio of nuclear strain to membrane strain to compensate for small variations in applied membrane strain. Cells that were damaged during or after strain application were excluded from the analysis.
Mouse Phenotypes
Disease phenotypes associated with Zmpste24 deficiency (abnormal body weight curves, reduced survival, and number of spontaneous rib fractures) were assessed in Zmpste24−/−Lmna+/+ and Zmpste24−/−LmnaLAO/+ mice, along with sibling control mice (Zmpste24+/+Lmna+/+, Zmpste24+/+LmnaLAO/+). Body weights were measured weekly for 20 weeks; numbers of surviving mice were also recorded weekly. The numbers of rib fractures were counted after removing the heart and lungs from the thoracic cavity (38, 41, 42).
Statistical Analyses
Statistical analyses were performed with Prism software version 5.0 (GraphPad). Statistical differences in the numbers of nuclei with nuclear blebs were assessed with the χ2 test. Variations in nuclear strain assays were analyzed with an unpaired t test with Welch's correction to account for differences in variances. Body weight curves were compared with repeated-measures ANOVA. Differences in the numbers of rib fractures were calculated with a two-tailed Student's t test. Survival differences were assessed by the Kaplan-Meier method using the log-rank test (41, 42).
RESULTS
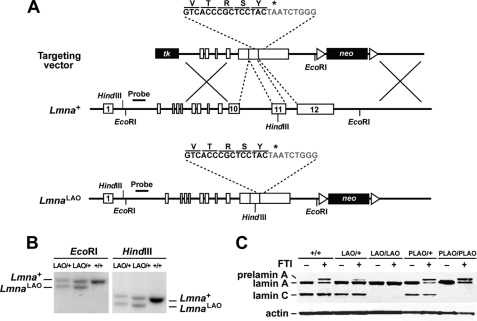
In the current study, we used gene targeting to create a mutant Lmna allele, LmnaLAO, that yields exclusively mature lamin A. The targeting event eliminated intron 10, thereby abolishing lamin C synthesis, and also deleted all DNA sequences between the last codon of mature lamin A (in exon 11) and the prelamin A stop codon (in exon 12) (Fig. 1A). Gene-targeting events were identified by Southern blotting of EcoRI- or HindIII-digested genomic DNA (Fig. 1B).
FIGURE 1.
Generation of the LmnaLAO allele. A, gene-targeting strategy to create a mutant Lmna allele, LmnaLAO, that yields exclusively mature lamin A. After recombination, introns 10 and 11 are deleted, thereby abolishing the synthesis of lamin C. Also, the last 30 bp of exon 11 and the first 24 bp of exon 12 are removed, placing the last codon within mature lamin A adjacent to the prelamin A stop codon (asterisk). Exon 12 sequences are indicated in gray. Exons are depicted as boxes; the position of the 5′-flanking probe and relevant restriction sites are also included. B, Southern blot identification of targeting events in embryonic stem (ES) cell clones with a 5′-flanking probe. Genomic DNA was digested with EcoRI or HindIII. EcoRI digestion yields a 10.4-kb wild-type band and a 9.4-kb band from the targeted allele (LmnaLAO); HindIII digestion yields a 8.5-kb wild-type band and a 7.8-kb LmnaLAO band. C, Western blots on extracts of mouse embryonic fibroblasts, grown either in the absence or the presence of an FTI (ABT-100, 5 μm). Lamin A, lamin C, and prelamin A were detected with a rabbit polyclonal antibody against lamin A/C. Actin was used as a loading control. The FTI resulted in the accumulation of prelamin A in all cells except LmnaLAO/LAO cells.
We predicted that the LmnaLAO allele would yield exclusively mature lamin A. To test this prediction, we performed Western blot analyses on extracts of LmnaLAO/+ and LmnaLAO/LAO embryonic fibroblasts with an antibody against lamin A/C. As controls, we examined fibroblasts from wild-type (Lmna+/+) embryos and embryos harboring a prelamin A-only allele (LmnaPLAO/+ and LmnaPLAO/PLAO). The LmnaPLAO allele contains a deletion of intron 10 (which abolishes lamin C synthesis) but no other mutations (43). As expected, LmnaLAO/LAO and LmnaPLAO/PLAO fibroblasts contained mature lamin A, but no lamin C (Fig. 1C). In LmnaLAO/LAO fibroblasts, lamin A was synthesized directly, whereas in LmnaPLAO/PLAO fibroblasts, it was generated through prelamin A processing. Thus, when prelamin A processing was blocked with a protein farnesyltransferase inhibitor (FTI), prelamin A accumulated in LmnaPLAO/PLAO fibroblasts, but not in LmnaLAO/LAO fibroblasts (Fig. 1C).
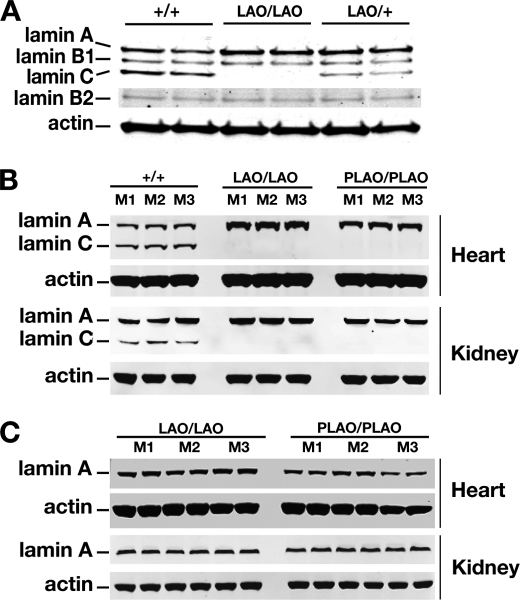
The amount of lamin A in primary LmnaLAO/LAO fibroblasts was greater than in Lmna+/+ fibroblasts (Fig. 2A). This result was not surprising, given that all of the output of the LmnaLAO allele is channeled into the production of mature lamin A (rather than split between lamin A and lamin C). The amounts of lamin B1 and B2 in LmnaLAO/LAO fibroblasts were similar to those in wild-type cells (Fig. 2A).
FIGURE 2.
Western blot analysis of lamin A and lamin C expression in extracts from fibroblasts and mouse tissues. A, Western blots of extracts from Lmna+/+, LmnaLAO/LAO, and LmnaLAO/+ fibroblasts (two cell lines/genotype) with antibodies against lamin A/C, lamin B1, lamin B2, and actin. B–C, Western blots of heart and kidney extracts from Lmna+/+, LmnaLAO/LAO, and LmnaPLAO/PLAO mice (three mice/genotype) with antibodies against lamin A/C (panel B), mature lamin A (panel C), and actin.
We observed consistent findings in heart and kidney extracts from LmnaLAO/LAO mice. The amount of mature lamin A in tissue extracts from LmnaLAO/LAO mice was greater than in extracts from Lmna+/+ mice (Fig. 2, B and C). Also, the amounts of mature lamin A in the heart and kidney of LmnaLAO/LAO and LmnaPLAO/PLAO mice were virtually identical (Fig. 2, B and C). Thus, eliminating the prelamin A processing pathway has little impact on the steady-state levels of mature lamin A in mouse tissues.
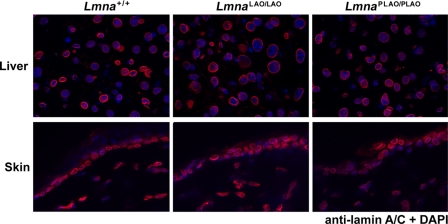
Earlier studies with transfected cells concluded that direct synthesis of mature lamin A substantially interferes with its ability to reach the nuclear envelope (19, 20). These studies led us to predict that LmnaLAO/LAO mice might exhibit disease phenotypes similar to those observed in Lmna knock-out mice (24) and that lamin A in LmnaLAO/LAO tissues might be mislocalized away from the nuclear envelope. Neither of these predictions was borne out. LmnaLAO/LAO mice were fertile and healthy, and the mice survived quite normally for more than two years. Body weights were entirely normal (supplemental Fig. S1), and routine histopathological studies did not uncover any abnormalities in any tissues, including skin, heart, brain, liver, skeletal muscle, and kidney (supplemental Fig. S2). Furthermore, the lamin A in the skin and liver of LmnaLAO/LAO mice was located largely at the nuclear rim; indistinguishable from Lmna+/+ and LmnaPLAO/PLAO mice (Fig. 3).
FIGURE 3.
Immunofluorescence microscopy showing that mature lamin A in tissues of LmnaLAO/LAO mice is located at the nuclear rim. Frozen sections of liver (top) and skin (bottom) of Lmna+/+, LmnaLAO/LAO, and LmnaPLAO/PLAO mice were stained with a lamin A/C antibody (red); nuclei were counterstained with DAPI (blue). Images were taken with a ×63 objective.
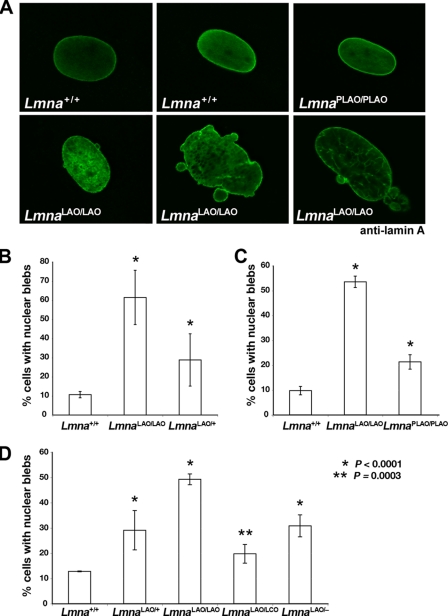
Whereas the nuclei in LmnaLAO/LAO tissues appeared normal, morphological abnormalities were detected in LmnaLAO/LAO fibroblasts. LmnaLAO/LAO fibroblasts had a higher frequency of nuclei with blebs (Fig. 4A). In three independent experiments with early-passage primary fibroblasts, the frequency of nuclear blebs in LmnaLAO/LAO fibroblasts was higher than in Lmna+/+ fibroblasts (p < 0.0001) (Fig. 4, B–D). LmnaPLAO/PLAO cells exhibited more blebbing than Lmna+/+ fibroblasts, but less than LmnaLAO/LAO fibroblasts (Fig. 4C). Cells containing a single LmnaLAO allele (LmnaLAO/+, LmnaLAO/LCO, LmnaLAO/−) also manifested a higher frequency of misshapen nuclei (p < 0.0001, except for the comparison of Lmna+/+ and LmnaLAO/LCO cells, where the p value was 0.0003) (Fig. 4, B and D).
FIGURE 4.
Misshapen cell nuclei in embryonic fibroblasts carrying either one or two LmnaLAO alleles. A, confocal microscopy images showing the presence of misshapen nuclei in LmnaLAO/LAO cells (stained with a lamin A antibody). Typical nuclei from Lmna+/+ and LmnaPLAO/PLAO fibroblasts are shown for comparison. B–D, three independent experiments assessing the frequency of nuclear blebs in primary embryonic fibroblasts from mice expressing one or two LmnaLAO alleles. B, comparison of the number of nuclear blebs in Lmna+/+ (n = 4), LmnaLAO/LAO (n = 2), and LmnaLAO/+ (n = 2) fibroblast cell lines, all isolated from sibling embryos. C, comparison of the number of nuclear blebs in primary Lmna+/+ (n = 2), LmnaLAO/LAO (n = 2), and LmnaPLAO/PLAO (n = 3) fibroblast cell lines. D, comparison of the frequency of nuclear blebs in primary Lmna+/+, LmnaLAO/+, LmnaLAO/LAO, LmnaLAO/LCO, and LmnaLAO/− fibroblasts (two cell lines/genotype). For each experiment, the frequency of nuclear blebs was assessed in >800 cells/genotype by observers blinded to genotype, as described (38, 41, 42). In each experiment, the frequency of nuclear blebs in LmnaLAO/LAO cells was higher than in Lmna+/+ cells (p < 0.0001). Cells containing one LmnaLAO allele (LmnaLAO/+, LmnaLAO/LCO, LmnaLAO/−) also had a higher frequency of misshapen nuclei than Lmna+/+ cells (p < 0.0001), except for the comparison between Lmna+/+ and LmnaLAO/LCO cells, where the p value was very slightly less (p = 0.0003).
As judged by confocal microscopy, some of the lamin A in LmnaLAO/LAO fibroblasts reached the nuclear rim, but the amount of lamin A at the nuclear rim (versus the amount in the nucleoplasm) appeared lower than in Lmna+/+ or LmnaPLAO/PLAO fibroblasts (Fig. 4A). We also imaged the nuclei of LmnaLAO/LAO fibroblasts with a Stimulated Emission Depletion confocal microscope, which provided higher-resolution images of the nuclear blebs (supplemental Fig. S3). A honeycomb-like pattern of staining was observed for both lamin A and Lap2β in the blebs of LmnaLAO/LAO fibroblasts. Fewer and smaller blebs were observed in Lmna+/+ fibroblasts, and they had a more homogenous pattern of staining for both proteins (Fig. S3).
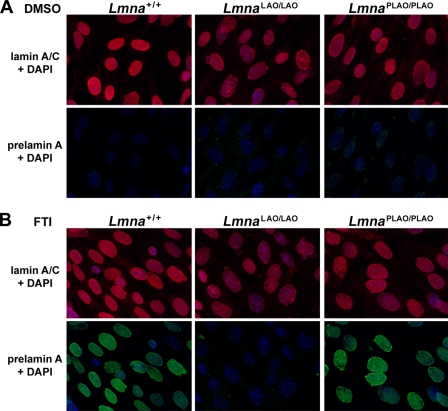
When Lmna+/+ and LmnaPLAO/PLAO fibroblasts were treated with an FTI, prelamin A accumulated and was easily detectable by immunofluorescence microscopy (Fig. 5). In contrast, prelamin A was absent in FTI-treated LmnaLAO/LAO fibroblasts (Fig. 5). We considered the possibility that the inability to synthesize prelamin A might interfere with the ability of lamin B1 and lamin B2 to reach the nuclear rim. However, this was not the case; both lamin B1 and lamin B2 reached the nuclear rim in LmnaLAO/LAO fibroblasts, indistinguishable from findings in Lmna+/+ fibroblasts (supplemental Fig. S4).
FIGURE 5.
Immunofluorescence microscopy of Lmna+/+, LmnaLAO/LAO, and LmnaPLAO/PLAO fibroblasts with an antibody against lamin A/C (red) and a rat monoclonal antibody against prelamin A (green). These images reveal a higher frequency of misshapen nuclei in LmnaLAO/LAO fibroblasts. Cells were grown in the presence of vehicle (DMSO) alone (panel A), or with an FTI (ABT-100, 2 μm) (panel B). Treatment of cells with an FTI results in an accumulation of prelamin A in Lmna+/+ and LmnaPLAO/PLAO fibroblasts, but not in LmnaLAO/LAO fibroblasts.
Because nuclear shape abnormalities can be indicative of impaired nuclear mechanics (25), we examined nuclear stiffness in LmnaLAO/LAO and LmnaPLAO/PLAO fibroblasts as well as Lmna+/+ control cells. LmnaPLAO/PLAO cells displayed decreased nuclear deformations under strain, suggesting increased nuclear stiffness, but nuclear stiffness in LmnaLAO/LAO and Lmna+/+ fibroblasts was indistinguishable (supplemental Fig. S5A). Also, a detailed comparison of nuclear stiffness in LmnaLAO/LAO cells with and without nuclear blebs (“abnormal” and “normal,” respectively) did not reveal any differences in nuclear stiffness between the two groups (supplemental Fig. S5B).
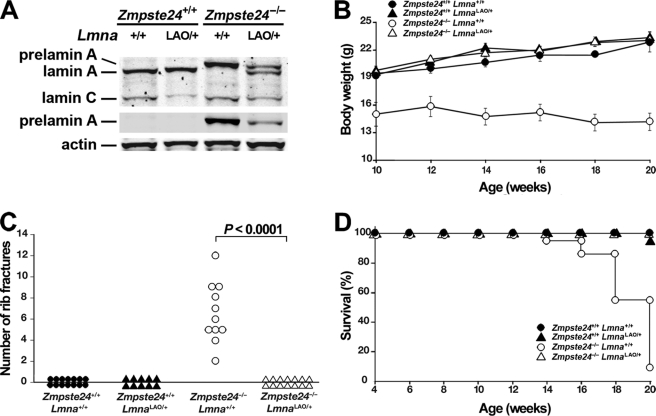
The accumulation of prelamin A in ZMPSTE24-deficient (Zmpste24−/−) mice leads to severe disease phenotypes (reduced body weight, spontaneous rib fractures, and reduced survival) (14, 15). We suspected that a single LmnaLAO allele, by reducing the production of prelamin A, would significantly reduce disease in Zmpste24−/− mice. Indeed, Zmpste24−/−LmnaLAO/+ mice produced less prelamin A than Zmpste24−/−Lmna+/+ mice and were entirely protected from the disease phenotypes that occur in Zmpste24−/−Lmna+/+ mice (Fig. 6).
FIGURE 6.
A single LmnaLAO allele eliminates disease phenotypes in Zmpste24−/− mice. A, Western blots of liver extracts with antibodies against lamin A/C, prelamin A, and actin. B–D, disease phenotypes in Zmpste24+/+Lmna+/+, Zmpste24−/−Lmna+/+, Zmpste24+/+LmnaLAO/+, and Zmpste24−/−LmnaLAO/+ mice. B, body weight curves of female mice. Zmpste24+/+Lmna+/+, n = 14; Zmpste24−/−Lmna+/+, n = 14; Zmpste24+/+LmnaLAO/+, n = 12; Zmpste24−/−LmnaLAO/+, n = 18. C, number of spontaneous rib fractures at the time of death (or when the experiment was terminated at 20 weeks). Zmpste24+/+Lmna+/+, n = 14; Zmpste24+/+LmnaLAO/+, n = 10; Zmpste24−/−Lmna+/+, n = 11; Zmpste24−/−LmnaLAO/+, n = 14. D, survival of male and female mice over a 20-week time span. The reduced survival phenotype in Zmpste24−/−Lmna+/+ mice was not observed in Zmpste24−/−LmnaLAO/+ mice (p < 0.0001). Zmpste24+/+Lmna+/+, n = 23; Zmpste24−/−Lmna+/+, n = 22; Zmpste24+/+LmnaLAO/+, n = 22; Zmpste24−/−LmnaLAO/+, n = 39.
DISCUSSION
The complex enzymatic pathway that converts prelamin A to mature lamin A is conserved in both avian and mammalian species (19, 21–23). Also, earlier transfection studies in cultured cells strongly suggested that prelamin A processing is required for the targeting of lamin A to the nuclear rim (19, 20). These findings led us to suspect that direct synthesis of lamin A, bypassing prelamin A synthesis and processing, would elicit severe disease phenotypes, perhaps similar to those in Lmna−/− mice (24). These suspicions were not confirmed. Mice that synthesize mature lamin A directly (LmnaLAO/LAO) exhibited no disease phenotypes or histopathological abnormalities, even after >24 months of observation. The remarkable health and vitality of LmnaLAO/LAO mice contrasts sharply with homozygous Lmna knock-out mice, which succumb to muscular dystrophy at 6 weeks of age (24), and with heterozygous Lmna knock-out mice, which manifest cardiomyopathy and conduction system disease after one year of age (26). The complete absence of disease in LmnaLAO/LAO mice led us to predict that the mature lamin A must reach the nuclear envelope normally, despite the absence of prelamin A synthesis and processing. Indeed, this was the case; the mature lamin A in tissues of LmnaLAO/LAO mice was positioned normally at the nuclear rim, indistinguishable from lamin A in wild-type and LmnaPLAO/PLAO mice. Also, we could not detect any peculiarities in nuclear shape in tissues of LmnaLAO/LAO mice.
Whereas no abnormalities in lamin A positioning were detectable in the tissues of LmnaLAO/LAO mice, we did observe a higher-than-normal frequency of misshapen nuclei in LmnaLAO/LAO fibroblasts. Although some of the lamin A clearly reached the nuclear rim in LmnaLAO/LAO fibroblasts, immunofluorescence microscopy pointed to increased amounts of mature lamin A in the nucleoplasm of those cells, compared with wild-type or LmnaPLAO/PLAO fibroblasts. The nucleoplasmic lamin A in LmnaLAO/LAO cells was often irregularly distributed, sometimes appearing in honeycomb-like lattices. Lamin A was also detectable in the nucleoplasm of wild-type or LmnaPLAO/PLAO fibroblasts, but in those cells, it was distributed homogenously. It is interesting that misshapen nuclei were also more frequent in cells harboring a single LmnaLAO allele. Thus, the nuclear shape abnormalities elicited by the mature lamin A from a LmnaLAO allele could not be prevented by prelamin A or lamin C synthesis from the other allele. We do not understand why alterations in nuclear morphology were obvious in fibroblasts while they were undetectable in tissues, but there are several possibilities. Fibroblasts grown on plastic plates are “spread out” and have a disk-shaped nucleus. In our studies, the thickness of a fibroblast nucleus is only 2–4 μm, compared with a cross-sectional diameter of 10–20 μm for nuclei in tissues. It is possible that the “flattening” of nuclei in cultured fibroblasts exaggerates nuclear lamina defects and nuclear shape abnormalities, making them easier to detect. Another possibility is that the abnormal distribution of lamin A and the accompanying nuclear shape abnormalities “snowball” in cell culture where the cells are growing continuously, perhaps giving cells less time for proper assembly of the lamina after each mitosis.
Interestingly, despite significant nuclear shape abnormalities, LmnaLAO/LAO fibroblasts had normal nuclear mechanics. These findings are reminiscent of lamin B1-deficient fibroblasts, which had severe nuclear blebbing but normal nuclear stiffness (25). We suspect that local defects in lamina organization may be sufficient to cause nuclear blebs, and can do so without affecting global nuclear stiffness.
Most cell transfection studies have pointed to a critical role of protein farnesylation in the targeting of lamin A to the nuclear rim (18–20). For example, Lutz et al. (19) reported that nonfarnesylated prelamin A is retained in the nucleoplasm and cannot reach the nuclear rim. On the other hand, Lutz et al. (19) found that a truncated prelamin A lacking the last 21 amino acids of the protein could reach the nuclear rim. Those observations prompted them to speculate that the last 21 amino acids of prelamin A might prevent prelamin A incorporation into higher order filaments in the nuclear lamina. They reasoned that prelamin A would become highly prone to polymerization only after cleavage by ZMPSTE24 at the nuclear rim. We believe that this speculation is intriguing. It is possible that the increased amount of lamin A in the nucleoplasm of LmnaLAO/LAO fibroblasts, along with its nonhomogenous distribution, could be due to “premature polymerization” of lamin A monomers (i.e. before the molecule reaches the nuclear rim).
Nuclear shape abnormalities have been identified in fibroblasts harvested from human muscular dystrophy patients with LMNA mutations (27) and the abnormalities in the nuclear lamina underlying these shape changes are often considered relevant to the pathogenesis of disease (28). For example, the misshapen nuclei in Lmna−/− fibroblasts (24) were accompanied by mechanical and signal transduction abnormalities that seemed highly relevant to disease (29). However, our current studies strongly suggest that misshapen nuclei in cultured fibroblasts do not necessarily foretell significant pathology in tissues. One might conceivably argue that the “disconnect” that we observed between misshapen nuclei in fibroblasts and disease phenotypes in mice might be unique to LmnaLAO/LAO mice, where an entirely normal nuclear lamin is produced in normal amounts. However, recent data suggests that the same “disconnect” can exist with structurally abnormal lamins. Muchir et al. (28) found no clear relationship between severity of disease in humans with LMNA mutations and morphological defects in the nuclei of cultured fibroblasts. Also, Wang et al. (30) generated transgenic mice that expressed progerin (the mutant prelamin A in Hutchinson-Gilford progeria syndrome) in skin keratinocytes. No skin pathology was detected in the mice, but keratinocytes exhibited nuclear shape alterations.
Genetic studies in mice suggested that a reduction in the synthesis of farnesyl-prelamin A reduces the progeria-like disease phenotypes in Zmpste24−/− mice (31). Our current studies provide further support for this concept. Zmpste24−/− mice carrying a single LmnaLAO allele accumulated less prelamin A than Zmpste24−/− mice harboring two wild-type Lmna alleles. Also, the single LmnaLAO allele, which reduces prelamin A production by 50%, completely abolished the disease phenotypes that normally occur in Zmpste24−/− mice.
A major finding of these studies is that LmnaLAO/LAO mice were healthy despite a complete absence of lamin C. Normally, mammalian cells and tissues express roughly equal amounts of lamin A and lamin C. Our studies show that lamin C synthesis is not essential for the health and vitality of the laboratory mouse. They also show that the lethal muscular dystrophy phenotype in Lmna-deficient mice (24) cannot be ascribed to the loss of lamin C.
In summary, the current studies provide two important lessons regarding lamin A biology. The first lesson is that direct synthesis of mature lamin A, bypassing prelamin A synthesis and processing, does not lead to detectable disease phenotypes. Misshapen nuclei were found in LmnaLAO/LAO fibroblasts, but no histopathology was detected in LmnaLAO/LAO mice, and remarkably, the targeting of mature lamin A to the nuclear rim appeared quite normal in tissues of LmnaLAO/LAO mice. Thus, prelamin A processing has minimal importance for the targeting of mature lamin A to the nuclear rim in mouse tissues. The second lesson is that the synthesis of lamin C is dispensable in the laboratory mouse.3
Supplementary Material
Acknowledgments
We thank David Frost from Abbott Pharmaceuticals for ABT-100. Confocal laser scanning microscopy was performed at the California NanoSystems Institute (CNSI) Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA, supported with funding from National Institutes of Health-NCRR shared resources Grant CJX1–44385-WS-29646 and NSF Major Research Instrumentation Grant CHE-0722519.
This work was supported, in whole or in part, by National Institutes of Health Grants AG035626, HL76839, HL86683, GM66152, HL082792, and NS059348. This work was also supported by March of Dimes Grant 6-FY2007-1012, the Ellison Medical Foundation Senior Scholar Program, two American Heart Association Scientist Development Grants 0835489N and 0635359N, and a postdoctoral fellowship award from the American Heart Association, Western States Affiliate.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
By saying that the prelamin A post-translational processing pathway is “dispensable,” we do not mean to suggest that prelamin A processing has been devoid of functional significance during vertebrate evolution. Indeed, we think that the conservation of this pathway means that it has had functional relevance. All we are saying is that prelamin A processing does not appear to be crucial for the overt health of the laboratory mice, or for the delivery of mature lamin A to the nuclear rim in tissues.
- ER
- endoplasmic reticulum
- STED
- stimulated emission depletion
- LAO
- lamin A-only
- PLAO
- prelamin A-only
- FTI
- protein farnesyltransferase inhibitor
- DAPI
- 4′,6-diamidino-2-phenylindole
- HIV
- human immunodeficiency virus.
REFERENCES
- 1.Sinensky M., Fantle K., Trujillo M., McLain T., Kupfer A., Dalton M. (1994) J. Cell Sci. 107, 61–67 [DOI] [PubMed] [Google Scholar]
- 2.Young S. G., Fong L. G., Michaelis S. (2005) J. Lipid Res. 46, 2531–2558 [DOI] [PubMed] [Google Scholar]
- 3.Corrigan D. P., Kuszczak D., Rusinol A. E., Thewke D. P., Hrycyna C. A., Michaelis S., Sinensky M. S. (2005) Biochem. J. 387, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young S. G., Ambroziak P., Kim E., Clarke S. (2000) in The Enzymes (Tamanoi F., Sigman D. S. eds), Vol. 21, pp. 155–213, Academic Press, San Diego, CA [Google Scholar]
- 5.Lin F., Worman H. J. (1993) J. Biol. Chem. 268, 16321–16326 [PubMed] [Google Scholar]
- 6.Worman H. J., Fong L. G., Muchir A., Young S. G. (2009) J. Clin. Invest. 119, 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriksson M., Brown W. T., Gordon L. B., Glynn M. W., Singer J., Scott L., Erdos M. R., Robbins C. M., Moses T. Y., Berglund P., Dutra A., Pak E., Durkin S., Csoka A. B., Boehnke M., Glover T. W., Collins F. S. (2003) Nature 423, 293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C. L., Munnich A., Le Merrer M., Lévy N. (2003) Science 300, 2055. [DOI] [PubMed] [Google Scholar]
- 9.Caron M., Auclair M., Sterlingot H., Kornprobst M., Capeau J. (2003) AIDS 17, 2437–2444 [DOI] [PubMed] [Google Scholar]
- 10.Coffinier C., Hudon S. E., Farber E. A., Chang S. Y., Hrycyna C. A., Young S. G., Fong L. G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13432–13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varela I., Pereira S., Ugalde A. P., Navarro C. L., Suárez M. F., Cau P., Cadiñanos J., Osorio F. G., Foray N., Cobo J., de Carlos F., Lévy N., Freije J. M., López-Otín C. (2008) Nat. Med. 14, 767–772 [DOI] [PubMed] [Google Scholar]
- 12.Navarro C. L., Cadiñanos J., De Sandre-Giovannoli A., Bernard R., Courrier S., Boccaccio I., Boyer A., Kleijer W. J., Wagner A., Giuliano F., Beemer F. A., Freije J. M., Cau P., Hennekam R. C., López-Otín C., Badens C., Lévy N. (2005) Hum. Mol. Genet. 14, 1503–1513 [DOI] [PubMed] [Google Scholar]
- 13.Moulson C. L., Go G., Gardner J. M., van der Wal A. C., Smitt J. H., van Hagen J. M., Miner J. H. (2005) J. Invest. Dermatol. 125, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergo M. O., Gavino B., Ross J., Schmidt W. K., Hong C., Kendall L. V., Mohr A., Meta M., Genant H., Jiang Y., Wisner E. R., Van, Bruggen N., Carano R. A., Michaelis S., Griffey S. M., Young S. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13049–13054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pendás A. M., Zhou Z., Cadiñanos J., Freije J. M., Wang J., Hultenby K., Astudillo A., Wernerson A., Rodríguez F., Tryggvason K., López-Otín C. (2002) Nat. Genet. 31, 94–99 [DOI] [PubMed] [Google Scholar]
- 16.Rusiñol A. E., Sinensky M. S. (2006) J. Cell Sci. 119, 3265–3272 [DOI] [PubMed] [Google Scholar]
- 17.Davies B. S., Fong L. G., Yang S. H., Coffinier C., Young S. G. (2009) Annu. Rev. Genomics Hum. Genet. 10, 153–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holtz D., Tanaka R. A., Hartwig J., McKeon F. (1989) Cell 59, 969–977 [DOI] [PubMed] [Google Scholar]
- 19.Lutz R. J., Trujillo M. A., Denham K. S., Wenger L., Sinensky M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3000–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennekes H., Nigg E. A. (1994) J. Cell Sci. 107, 1019–1029 [DOI] [PubMed] [Google Scholar]
- 21.Vorburger K., Kitten G. T., Nigg E. A. (1989) EMBO J. 8, 4007–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber K., Plessmann U., Traub P. (1989) FEBS Lett. 257, 411–414 [DOI] [PubMed] [Google Scholar]
- 23.Beck L. A., Hosick T. J., Sinensky M. (1990) J. Cell Biol. 110, 1489–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C. L., Burke B. (1999) J. Cell Biol. 147, 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lammerding J., Fong L. G., Ji J. Y., Reue K., Stewart C. L., Young S. G., Lee R. T. (2006) J. Biol. Chem. 281, 25768–25780 [DOI] [PubMed] [Google Scholar]
- 26.Wolf C. M., Wang L., Alcalai R., Pizard A., Burgon P. G., Ahmad F., Sherwood M., Branco D. M., Wakimoto H., Fishman G. I., See V., Stewart C. L., Conner D. A., Berul C. I., Seidman C. E., Seidman J. G. (2008) J. Mol. Cell Cardiol. 44, 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonne G., Di Barletta M. R., Varnous S., Bécane H. M., Hammouda E. H., Merlini L., Muntoni F., Greenberg C. R., Gary F., Urtizberea J. A., Duboc D., Fardeau M., Toniolo D., Schwartz K. (1999) Nat. Genet. 21, 285–288 [DOI] [PubMed] [Google Scholar]
- 28.Muchir A., Medioni J., Laluc M., Massart C., Arimura T., van der Kooi A. J., Desguerre I., Mayer M., Ferrer X., Briault S., Hirano M., Worman H. J., Mallet A., Wehnert M., Schwartz K., Bonne G. (2004) Muscle Nerve 30, 444–450 [DOI] [PubMed] [Google Scholar]
- 29.Lammerding J., Schulze P. C., Takahashi T., Kozlov S., Sullivan T., Kamm R. D., Stewart C. L., Lee R. T. (2004) J. Clin. Invest. 113, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Panteleyev A. A., Owens D. M., Djabali K., Stewart C. L., Worman H. J. (2008) Hum. Mol. Genet. 17, 2357–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong L. G., Ng J. K., Lammerding J., Vickers T. A., Meta M., Coté N., Gavino B., Qiao X., Chang S. Y., Young S. R., Yang S. H., Stewart C. L., Lee R. T., Bennett C. F., Bergo M. O., Young S. G. (2006) J. Clin. Invest. 116, 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S. H., Bergo M. O., Toth J. I., Qiao X., Hu Y., Sandoval S., Meta M., Bendale P., Gelb M. H., Young S. G., Fong L. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10291–10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joyner A. L., Skarnes W. C., Rossant J. (1989) Nature 338, 153–156 [DOI] [PubMed] [Google Scholar]
- 34.Leung G. K., Schmidt W. K., Bergo M. O., Gavino B., Wong D. H., Tam A., Ashby M. N., Michaelis S., Young S. G. (2001) J. Biol. Chem. 276, 29051–29058 [DOI] [PubMed] [Google Scholar]
- 35.Kim E., Ambroziak P., Otto J. C., Taylor B., Ashby M., Shannon K., Casey P. J., Young S. G. (1999) J. Biol. Chem. 274, 8383–8390 [DOI] [PubMed] [Google Scholar]
- 36.Steinert P., Zackroff R., Aynardi-Whitman M., Goldman R. D. (1982) Methods Cell Biol. 24, 399–419 [DOI] [PubMed] [Google Scholar]
- 37.Lee R., Chang S. Y., Trinh H., Tu Y., White A. C., Davies B. S., Bergo M. O., Fong L. G., Lowry W. E., Young S. G. (2010) Hum. Mol. Genet. 19, 1603–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fong L. G., Ng J. K., Meta M., Coté N., Yang S. H., Stewart C. L., Sullivan T., Burghardt A., Majumdar S., Reue K., Bergo M. O., Young S. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 18111–18116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toth J. I., Yang S. H., Qiao X., Beigneux A. P., Gelb M. H., Moulson C. L., Miner J. H., Young S. G., Fong L. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12873–12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang S. H., Qiao X., Farber E., Chang S. Y., Fong L. G., Young S. G. (2008) J. Biol. Chem. 283, 7094–7099 [DOI] [PubMed] [Google Scholar]
- 41.Yang S. H., Meta M., Qiao X., Frost D., Bauch J., Coffinier C., Majumdar S., Bergo M. O., Young S. G., Fong L. G. (2006) J. Clin. Invest. 116, 2115–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fong L. G., Frost D., Meta M., Qiao X., Yang S. H., Coffinier C., Young S. G. (2006) Science 311, 1621–1623 [DOI] [PubMed] [Google Scholar]
- 43.Davies B. S., Barnes R. H., Tu Y., Ren S., Andres D. A., Spielmann H. P., Lammerding J., Wang Y., Young S. G., Fong L. G. (2010) Hum. Mol. Genet., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.