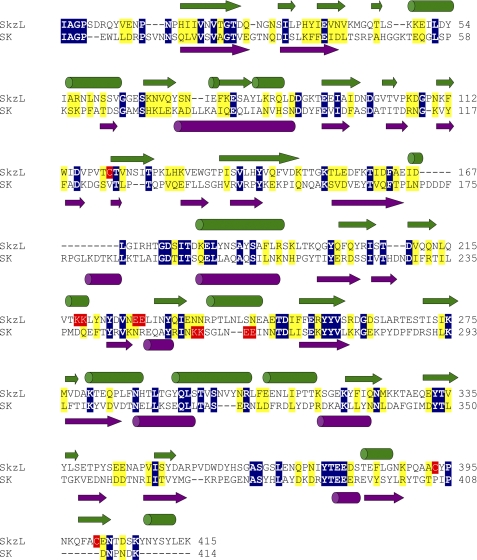
FIGURE 1.
Sequence alignment and secondary structure prediction for SkzL and SK. Sequence alignment of SkzL and SK from S. equisimilis was performed using the Clustal 2.0.10 multiple sequence alignment algorithm (69). Identical residue pairs are highlighted in blue with conserved residues in yellow. Secondary structure prediction was performed by PSIPRED for SkzL in green and SK in violet (68). SkzL Cys120, Cys 393, and Cys401 are highlighted in red. The pairs of Lys and Glu residues representing part (Lys256 and Lys257) of the SK motif involved in kringle 5-mediated Pg substrate recognition and the pairs of Lys and Glu residues in SkzL that may be involved in the low affinity LBS-dependent interaction of SkzL with Pg are also highlighted in red (42).