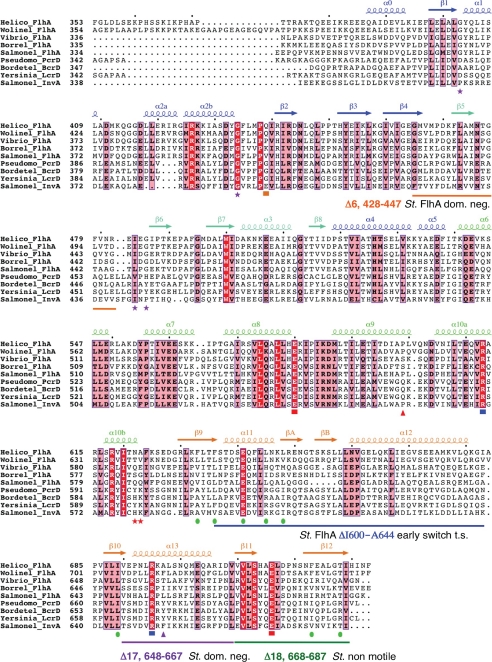
FIGURE 2.
Structurally based alignment of FlhAC and type III secretion homologue sequences. Calculated with T-Coffee (51) and rendered using Espript (52). Secondary structure assignments for the x-ray structure of H. pylori FlhAC are shown, and labeled according to the Salmonella FlhAC structure (23). Highly conserved residues are boxed in red, moderately conserved residues in pink. Salmonella temperature-sensitive secretion point mutations in FlhAC are marked with purple stars (19). Salmonella flhA nonsense mutations (Q588Stop, Q589Stop) that permit early secretion of FlgM are marked with red stars (48). The Salmonella flhA V404M mutation is marked with an orange square (13, 17). Y. pestis LcrD secretion-defective mutants are marked with triangles, red indicates a nonsense mutation at Gln-574 that deletes the C-terminal domain, pink depicts a Y670C missense mutation (34, 35). Conserved salt bridge interactions between the helical and C-domains of H. pylori FlhAC are marked with red (Glu) or blue (Arg) squares. Green circles mark residues that make up a predominantly hydrophobic surface on the C-domain. Salmonella FlhA deletion mutants mentioned in the text are marked by colored lines (16, 48).