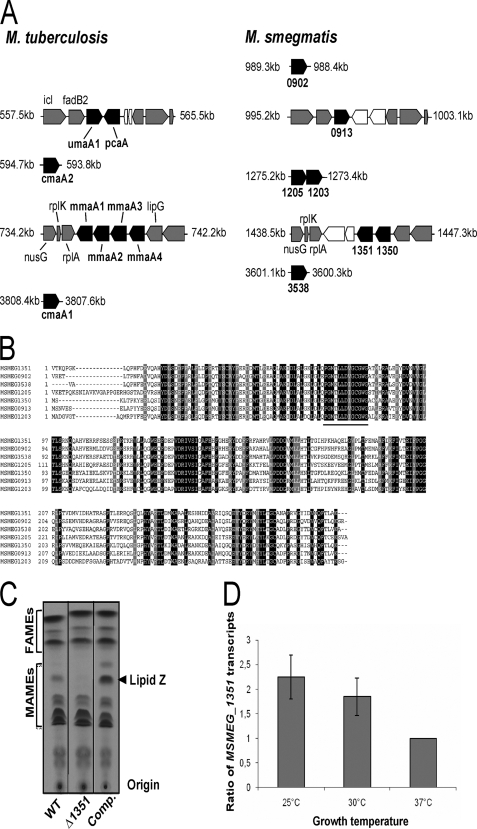
FIGURE 2.
Identification of the genes involved in the production of the cyclopropanated mycolates. A, schematic representation and genomic organization of the potential methyltransferases in M. smegmatis and comparison with M. tuberculosis. Black boxes indicate the mycolic acid methyltransferases; gray boxes indicate open reading frames encoding conserved proteins between the two species, and white boxes represent open reading frames unique to either M. smegmatis or M. tuberculosis. Coordinates of each gene cluster are indicated in kb. B, alignment of the M. smegmatis putative methyltransferases. The AdoMet-binding site as described previously (14) is underlined. C, argentation TLC of 14C-radiolabeled mycolic acids from the parental strain (1st lane), the ΔMSMEG_1351 mutant carrying either the empty pMV306 (2nd lane) or pMV306_1351 (3rd lane), all grown at 25 °C. The arrowhead indicates the position of lipid Z that is lacking in the ΔMSMEG_1351 mutant. D, temperature-regulated expression of MSMEG_1351. Wild-type M. smegmatis was grown at the indicated temperature, and the levels of MSMEG_1351 transcripts relative to those of the RNA polymerase σ-factor mysA gene (MSMEG_2758) were measured by quantitative reverse transcription-PCR. MSMEG_1351 transcript levels at the different temperatures were normalized to those from cells growing at 37 °C. Results are expressed as means ± S.D. from four independent experiments.