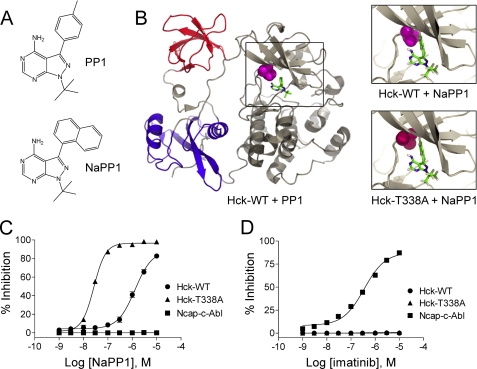
FIGURE 1.
Generation of NaPP1-sensitive Hck. A, structures of the non-selective SFK inhibitor PP1 and the bulky inactive analog, NaPP1 are shown. B, modeling of NaPP1 in the active site of wild-type Hck and the gatekeeper mutant, T338A is shown. The overall structure of Hck is shown on the left, with the SH3 domain in red, SH2 domain in blue, and the kinase domain in gray. The side chain of the gatekeeper residue (Thr-338) is highlighted in magenta, and its relationship to PP1 is shown in the boxed area. The model is based on the crystallographic coordinates of Schindler et al. (30) (PDB code 1QCF). Upper right, the spatial coordinates of the PP1 pyrazolo-pyrimidine were used to model the position of NaPP1 within the ATP binding site. This close-up view shows the clash of the naphthyl ring of NaPP1 with the side chain of the gatekeeper threonine. WT, wild type. Lower right, the T338A mutation was modeled in the Hck structure with the alanine side chain highlighted in red. This substitution creates a space that accommodates the naphthyl moiety of NaPP1 and sensitizes the kinase to this modified inhibitor. C and D, the Hck-T338A mutant is sensitive to NaPP1 but not to imatinib in an in vitro kinase assay. Recombinant wild-type Hck-YEEI (WT), Hck-T338A-YEEI, and Ncap-c-Abl were purified from Sf9 insect cells, and kinase activity was assessed in vitro using a fluorescence resonance energy transfer-based assay with a peptide substrate. Concentration-response curves are shown in the presence of NaPP1 (C) or imatinib (D). Percent inhibition is expressed as the mean ± S.D. from the results of four assay wells per condition. The entire experiment was repeated twice and produced comparable results; a representative example is shown.