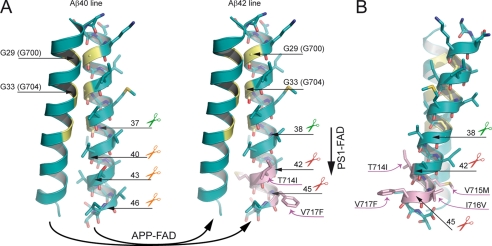
FIGURE 6.
Model of the pathological mechanisms caused by APP-FAD and PS1-FAD mutations. Oxygen atoms are depicted in red, nitrogen atoms are depicted in blue, and sulfur atoms are colored dark yellow. The glycine residues of the interface are highlighted in yellow, and APP-FAD mutations are colored pink. Scissors indicate peptide bonds that are cleaved by the γ-secretase. A, dimeric wt substrates are degraded by the γ-secretase predominantly by the Aβ40 line. Left, corresponding peptide bonds are indicated and are located at the dimer interface. We propose that APP-FAD mutations cause a general shift between the two product lines so that the Aβ40 line is down-regulated, and the Aβ42 line becomes a major degradation pathway. Right, APP-FAD, peptide bonds of the Aβ42 line are indicated. Note, that amino acid side chains from FAD mutations do not reach into the dimer interface. PS1-FAD mutants seem to cause substrate flux inhibition leading to a retarded processing within the Aβ42 line causing increased Aβ42 and decreased Aβ38 levels (vertical arrow). The dimer crossing point mediated by Gly29 (Gly700) and Gly33 (Gly704) may cause a steric hindrance and inhibit the consecutive γ-secretase processing leading predominantly to Aβ40 (Aβ40 line) or to Aβ42 (Aβ42 line) (9, 10). Mechanistically, the effect of G33A occurs after the effects of APP-FAD mutations explaining why G33A causes a constant reduction by 60% for Aβ42 and a 3-fold increase of Aβ38 in the presence of all individual APP-FAD mutations analyzed. B, the APP-FAD-TMS dimer in side view for better illustration of the FAD-causing amino acid side chains. T714I, V715M, I716V, and V717F stick out of the dimer interface and thus likely affect processing by modulating the substrate-enzyme recognition.