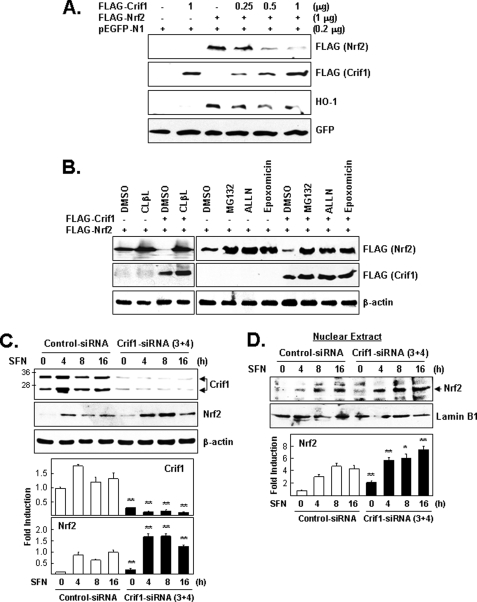
FIGURE 3.
CRIF1 regulates NRF2 protein levels via proteasome-mediated degradation. A, effect of CRIF1 overexpression on NRF2 protein levels. Cells (HEK293) were co-transfected for 24 h with three expression vectors (constant amounts of FLAG-NRF2 and pEGFP-N1 (the control vector) and increasing amounts of FLAG-CRIF1) and were analyzed on WB. B, effect of proteasomal inhibitors on CRIF1-driven down-regulation of NRF2 protein levels. Cells co-transfected as in A with the indicated expression vectors were incubated for an additional 4 h with proteasomal inhibitors (clasto-lactacystin β-lactone (CLβL) (10 μm), MG132 (10 μm), ALLN (50 μm), and epoxomicin (1 μm)) and then analyzed by WB. Exogenous FLAG-CRIF1 and FLAG-NRF2 were identified by their different migration distances. C, effects of CRIF1 knockdown on NRF2 protein levels. Cells (MCF-7) pretreated with siRNA (control versus CRIF1–3 and -4) for 72 h were treated with SFN (5 μm) and harvested at the indicated times. Total cell lysates were used for WB analysis with anti-NRF2 and anti-CRIF1 mouse monoclonal antibodies. β-Actin was used as a loading and transfer control. D, nuclear extracts from cells in C were subjected to WB analysis. Lamin B1 was used as a control for nuclear fractionation and equal loading. The results of WB image quantification of triplicate experiments are shown as bar graphs on the bottom panels of C and D.