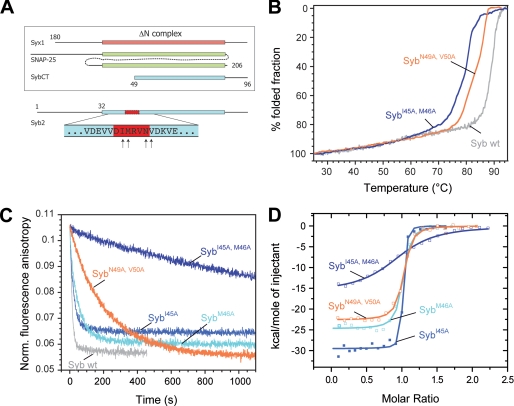
FIGURE 2.
Mutations in the putative coiled coil trigger site strongly inhibit binding of synaptobrevin 2 to the ΔN complex. A, depiction of the constructs used for binding experiments between the ΔN complex and synaptobrevin. The purified ΔN complex was composed of SyxH3 (aa 180–262), full-length SNAP-25a (aa 1–206), and Syb49–96. The arrows at the bottom of the diagram indicate the residues mutated in synaptobrevin (aa 1–96). B, thermal unfolding of SNARE complexes was monitored by CD spectroscopy at 222 nm. Purified SNARE complexes containing the double mutants SybI45A,M46A and SybN49A,V50A unfolded at slightly lower temperatures than the complex containing wild-type (wt) synaptobrevin (18). C, upon binding, the Alexa 488-labeled synaptobrevin fragment Syb49–96 is displaced from the ΔN complex (SyxH3·SNAP-25·Syb49–96C79Alexa488). As demonstrated before (12, 13), displacement is visible by a decrease in fluorescence anisotropy. Approximately 100 nm of the ΔN complex was incubated with different synaptobrevin mutants (500 nm). In agreement with our previous findings, the labeled fragment was quickly displaced upon addition of synaptobrevin (12, 13). Displacement was significantly slower when the double mutants SybI45A,M46A and SybN49A,V50A were used. D, isothermal titrations of different synaptobrevin mutants into the ΔN complex. Titration of SybI45A (18 μm), SybM46A (20 μm), and SybN49A,V50A (22 μm) into 2.5 μm purified ΔN complex (SyxH3·SNAP-25·Syb49–96); 38 μm SybI45A,M46A was titrated in 4 μm ΔN complex.