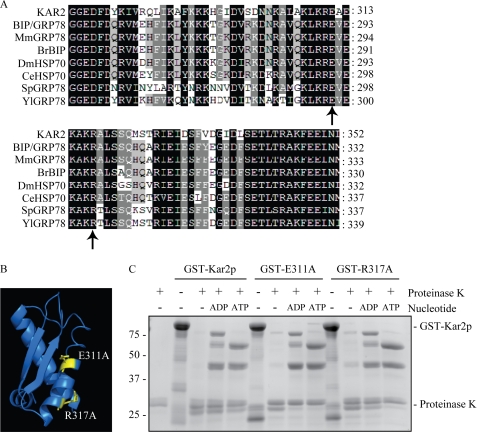
FIGURE 4.
Mutations in the IIB domain of Kar2p do not perturb the Kar2p nucleotide-dependent conformational change. A, sequence alignment of the IIB domain of Kar2p with other Kar2p homologues using ClustalW software. Sequences aligned are as follows: Mm, Mus musculus; Br, Brachydanio rerio; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sp, Schizosaccharomyces pombe, and Yl, Yarrowia lipolytica, and the residues numbers are annotated. Residues chosen for mutagenesis are indicated with a black arrow. Black shading represents 100% sequence identity; dark gray shading represents 80% sequence identity, and light gray shading represents 60% sequence identity. B, location of the residues chosen for mutagenesis in the IIB domain of the structural model of Kar2p. C, protease protection assay of wild-type and mutant Kar2p. 5 μg of protein was incubated with nucleotide for 30 min followed by 5 min of digestion with proteinase K. Digestion products were analyzed by Coomassie staining.