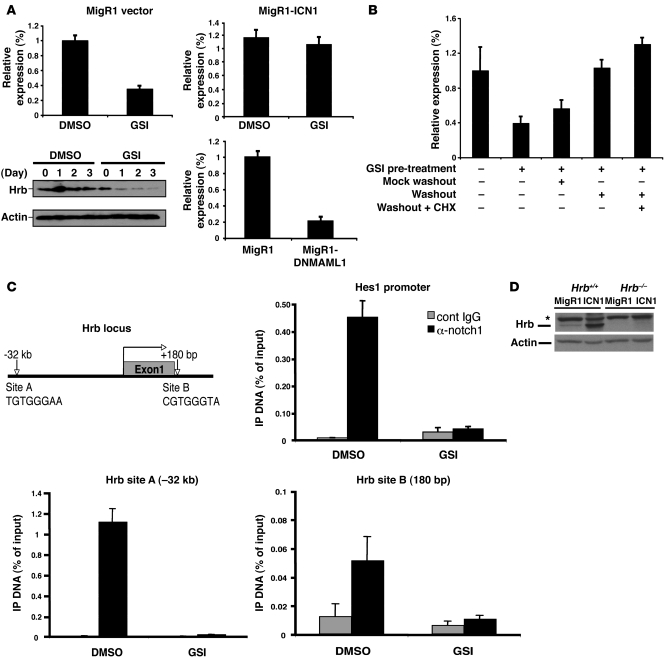
Figure 2. Hrb is a direct Notch1-regulated transcriptional target.
(A) A murine Notch-dependent cell line, T6E12, was utilized for Hrb mRNA and protein analysis. T6E12 cells were transduced with MigR1 (empty vector), MigR1-ICN1, or MigR1-DNMAML1 for 48 hours followed by 24 hours DMSO or GSI (1 μM) treatment as labeled. Hrb mRNA and protein levels were determined by quantitative RT-PCR and Western blot analysis, respectively. Results were obtained from 3 independent experiments. (B) T6E12 cells were treated with GSI (1 μM) for 48 hours to permit accumulation of the gamma-secretase substrate (transmembrane-Notch1 [NTM]). Cells were then washed and replenished with medium containing GSI (mock washout) or medium lacking GSI (washout) in the absence or presence of 20 μM cycloheximide (CHX). Hrb mRNA levels were evaluated by quantitative RT-PCR after 4 hours of additional incubation. Similar results were obtained in 3 independent experiments. (C) ChIP was performed on cross-linked fragmented DNAs prepared from T6E12 cells treated with DMSO or 1 μM GSI for 24 hours. Eluted DNAs were then analyzed by quantitative RT-PCR using primers flanking putative CSL-binding sites (A and B). Amplification of hes1 CSL-binding site served as a positive control to validate ChIP efficiency. The amount of DNA amplified from immunoprecipitated DNAs was normalized to that amplified from input DNA. (D) Western blot of lysates prepared from empty vector– (MigR1) and ICN1-transduced Hrb+/+ and Hrb–/– T cell precursors blotted for Hrb (clone H-300) and actin (loading control). Asterisk indicates nonspecific band. Data are shown as mean ± SD.