Nitrate assimilation genes in Chlamydomonas are negatively modulated by ammonium through a pathway that involves NO, cGMP, and calcium. This work shows that similar mechanisms might be operating in plants.
Abstract
Nitrate assimilation in plants and related organisms is a highly regulated and conserved pathway in which the enzyme nitrate reductase (NR) occupies a central position. Although some progress has been made in understanding the regulation of the protein, transcriptional regulation of the NR gene (NIA1) is poorly understood. This work describes a mechanism for the ammonium-mediated repression of NIA1. We report the characterization of a mutant defective in the repression of NIA1 and NR in response to ammonium and show that a gene (CYG56) coding for a nitric oxide (NO)-dependent guanylate cyclase (GC) was interrupted in this mutant. NO donors, cGMP analogs, a phosphodiesterase inhibitor isobutylmethylxanthine (IBMX), and a calcium ionophore (A23187) repress the expression of NIA1 in Chlamydomonas reinhardtii wild-type cells and also repress the expression of other ammonium-sensitive genes. In addition, the GC inhibitors LY83,583 (6-anilino-5,8-quinolinedione) and ODQ (1H-[1,2,4]oxadiazolo-[4,3-a]quinoxalin-1-one) release cells from ammonium repression. Intracellular NO and cGMP levels were increased in the presence of ammonium in wild-type cells. In the cyg56 mutant, NIA1 transcription was less sensitive to NO donors and A23187, but responded like the wild type to IBMX. Results presented here suggest that CYG56 participates in ammonium-mediated NIA1 repression through a pathway that involves NO, cGMP, and calcium and that similar mechanisms might be occurring in plants.
INTRODUCTION
Regulation of the nitrate assimilation pathway coordinates the incorporation of nitrogen with important processes of plant development, such as cell differentiation and photosynthesis. Genes and proteins of this pathway are induced by nitrate itself (Crawford, 1995; Daniel-Vedele et al., 1998; Forde, 2000; Llamas et al., 2002; Rexach et al., 2002) and are repressed by ammonium or by products of nitrate metabolism, such as Gln and Glu (Stitt, 1999; Crawford and Forde, 2002; Glass, 2003; Fan et al., 2006; Fernandez and Galván, 2007). A key component of the nitrate assimilation pathway is nitrate reductase (NR), which catalyzes the reduction of nitrate to nitrite. This enzyme and the gene encoding it are subject to complex regulation mechanisms: not only are gene expression and protein activity regulated by inorganic nitrogen compounds and metabolic derivatives, but they also respond to external cues, such as day and night cycles (Kaiser and Brendle-Behnisch, 1991; Galangau et al., 1998). In plants, NR activity is regulated by phosphorylation of a Ser residue that allows binding of 14-3-3 proteins and leads to subsequent degradation of NR (reviewed in Kaiser and Huber, 2001). Two kinases responsible for the phosphorylation of the Ser residue seem to be activated by calcium, whereas the third, a member of the SNF1 kinase family, is calcium independent (Douglas et al., 1997).
The green alga Chlamydomonas reinhardtii and higher plants share structural and regulatory elements for nitrate assimilation, making Chlamydomonas an interesting and useful model organism for studies of nitrogen metabolism (Galvan et al., 2006). The 14-3-3 proteins also appear to regulate nitrogen assimilation in Chlamydomonas; in fact, glutamine synthetase I is a target for these regulatory proteins (Pozuelo et al., 2001). However, redox regulation seems to be the major mechanism in regulating NR protein activity in the alga.
In Chlamydomonas, NIT2 is essential for nitrate assimilation by mediating transcriptional activation of NIA1, the gene encoding NR, in response to nitrate (Schnell and Lefebvre, 1993; Fernández et al., 1998; Camargo et al., 2007). This protein, containing GAF and RWP-RK domains, shows conservation with plant nodule inception-like proteins that might function similarly (Camargo et al., 2007; Castaigns et al., 2009). By contrast, the mechanism of negative regulation of NIA1 expression is poorly understood.
In a previous study, we reported the generation of a Chlamydomonas insertional mutant library whose objective was to identify new regulators of NIA1 expression (González-Ballester et al., 2005a). The genetic background of the parental strain 704 contained the NIA1 promoter fused to the arylsulfatase (Ars) reporter gene, so that NIA1 promoter activity was detected by ARS activity measurements in the conditions of the screen. A specific class of mutants, hereafter called ammonium insensitive (AI), was screened for NIA1 promoter activity in medium containing nitrate and ammonium, a condition in which NIA1 expression is not detected in the wild type. The tag was localized in the genome of the AI mutants, allowing us to identify a number of candidate genes involved in the repression of NIA1 (González-Ballester et al., 2005a, 2005b). One of these genes showed significant homology with soluble nitric oxide (NO)-dependent guanylate cyclases (GCs) from animals.
An exponentially growing number of reports on the implication of NO and cGMP in physiological processes are accumulating in plants. NO was first demonstrated to function as a signal molecule in plant–pathogen interactions (Delledonne et al., 1998), and it has subsequently been shown to play an active role in regulating root elongation (Gouvea et al., 1997), abscisic acid–induced stomatal closure (Mata and Lamattina, 2001), flowering He et al., 2004), senescence (Corpas et al., 2004), development of cell polarity (Salmi et al., 2007), and other processes. These functions have been extensively reviewed by Lamattina et al. (2003) who suggested the concept of NO functioning as a “synchronizing chemical messenger” to explain its implication in such diverse regulation pathways. In addition, cGMP signaling has been related to defense gene induction (Durner et al., 1998), cell polarity development (Salmi et al., 2007), or gene transcription and cation transport processes (Maathuis, 2006).
NR has been shown to synthesize NO using nitrite as a substrate (Dean and Harper, 1986, 1988; Yamasaki and Sakihama, 2000; Rockel et al., 2002; Sakihama et al., 2002), but little is known about the conditions regulating NO production in vivo. Mitochondrial electron transport was also found to contribute significantly to the nitrite-dependent NO production in plant cells (Planchet et al., 2005). In addition, nitrite is a known source of NO through nonenzymatic reactions (Bethke et al., 2004; Tischner et al., 2004). Interestingly, nitrite has been shown to inhibit transcription of NIA1 in Arabidopsis thaliana and Chlamydomonas (Llamas et al., 2002; Loqué et al., 2003). Nevertheless, evidence of NO as a regulator of nitrate assimilation is restricted to NR activity in tomato (Solanum lycopersicum) roots depending on levels of nitrate supply (Jin et al., 2009).
In this work, we characterize CYG56, a GC gene related to the AI phenotype, and show that NO, cGMP, and calcium participate in a signaling pathway for ammonium repression in Chlamydomonas. We also propose a model signaling pathway that might also be operative in plants.
RESULTS
A GC Is Related to Ammonium Repression of NIA1
AI insertional mutants have been selected for derepression of NIA1 expression using media containing nitrate and ammonium, and the location of the insertion tag in the genome of each of the 40 AI mutants was determined. This allowed us to identify candidate genes involved in negative regulation of NIA1 transcription (González-Ballester et al., 2005a, 2005b). In one of these mutants, previously known as mutant 20.73, a gene annotated as CYG56 (EU841916) was interrupted. Hereafter in the text, mutant 20.73 will therefore be referred to as cyg56. AI phenotype of this mutant was determined by ARS activity assays (Ohresser et al., 1997) after growth on solid medium containing different sources of nitrogen as previously described (González-Ballester et al., 2005a).
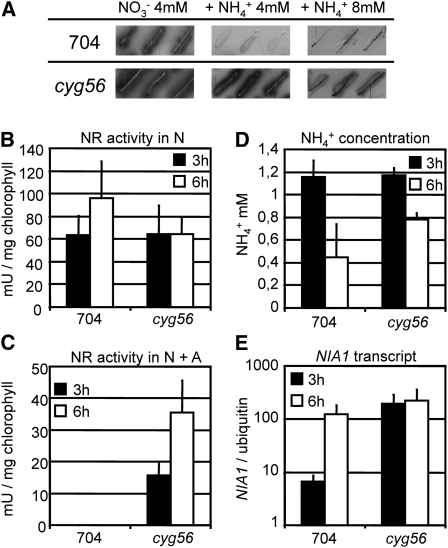
In contrast with the parental wild-type strain 704, the expression of the ARS reporter under control of the NIA1 promoter could be detected in cyg56 mutant cells in media containing nitrate plus ammonium, though the intensity of the signal decreased with increasing amounts of ammonium in the medium (Figure 1A). The phenotype was confirmed in short-term NR activity assays in medium containing nitrate plus ammonium; NR activity could be detected in the mutant but not in the wild type (Figure 1C). In these conditions, NR activity in the mutant increased with time as ammonium concentration, still present, decreased (Figure 1D). Importantly, there was no change in NR activity between the strains in nitrate medium alone (Figure 1B).
Figure 1.
NIA1 Expression and NR Activity Are Derepressed in the cyg56 Mutant.
(A) ARS activity of Chlamydomonas wild-type strain 704 and mutant cyg56 was assayed after 4 d of growth on solid media containing 4 mM NO3−4 mM NO3− + 4 mM NH4+, and 4 mM NO3− + 8 mM NH4+. Both strains contain the Ars gene under the control of the NIA1 promoter in their genetic background, and ARS activity therefore reflects NIA1 promoter activity.
(B) and (C) NR activity of strains 704 and cyg56 was assayed in media 4 mM NO3− (B) and 4 mM NO3− + 2 mM NH4+ (C) after incubation for 3 h (black bars) and 6 h (white bars).
(D) Remaining concentration of NH4+ at both time points of results shown in (B) and (C), respectively.
(E) NIA1 mRNA levels were measured by real-time PCR in the same conditions as in (C). The error bars represent the sd of three biological replicates.
Taken together, these results show (1) that the phenotype was not due to general activation of NR activity but to a reduced sensitivity of cyg56 to ammonium, and (2) that cyg56 shows an AI phenotype for NR activity still sensitive to increasing amounts of ammonium, consistent with the results of Figure 1A.
NIA1 transcript levels (Figure 1E) were then determined in the same conditions than Figure 1C. In agreement with the previous data, results showed that NIA1 mRNA levels were higher in the mutant compared with the wild type at both time points, the difference being more pronounced after 3 h of induction. Detection of NIA1 expression in the wild type was not surprising as it brought to light previously described posttranscriptional negative effects of ammonium on NR activity (Fernández et al., 1998).
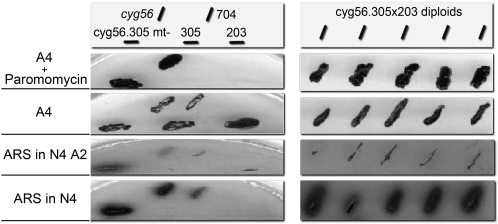
Linkage between marker insertion in the CYG56 gene and the AI phenotype in the mutant is supported by the following facts. First, a single insertion tag of the paromomycin marker was found in mutant cyg56 by DNA gel blot and PCR analyses (González-Ballester et al., 2005b). Second, genetic crosses between strain cyg56 paromomycin-resistant AI mt+ and different Chlamydomonas mt- strains that are paromomycin sensitive and sense normally ammonium were performed. Even though crossing efficiency was very low, as reported for these cell wall–less mutants (Galvan et al., 2007), progeny were obtained and analyzed. A linkage between paromomycin resistance and the ammonium-insensitive phenotype (determined from NIA1-ARS activity) was found in all 27 segregants recovered that had the chimeric NIA1-ARS reporter. In these segregants, the intensity of the AI phenotype ranged from strong to weak, indicating that genomic background of particular segregants might modulate this phenotype. Third, the cyg56 mutation was complemented by forming diploid cells. To do that, cyg56 mutation was transferred to a 305nia1- mutant background by genetic crosses. The transfer of the AI phenotype to NR mutant background also resulted in a strong desensitizing of the NIA1 reporter to ammonium (Figure 2, cyg56.305mt-), probably due to nitrate accumulation in the cells because of the lack of nitrate assimilation (Llamas et al., 2002). Then, Nit+ diploids (able to grow on nitrate) were recovered from mutant strains cyg56.305 (NIA1- and mt-) with strain 203 (Nit2- and mt+) by complementation. All diploids (cyg56.305x203) obtained that were paromomycin resistant had recovered the normal ammonium sensitivity of wild-type 704 as assayed by ARS activity (Figure 2).
Figure 2.
Complementation Analysis of the cyg56 Mutation.
The mutation cyg56 and the Ars marker were transferred to a 305nia1 mutant background. Nit+ diploid strains were isolated by in vivo complementation between 305nia1 and 203nit2 mutants and the AI phenotype of complemented diploids analyzed in medium with or without 4 mM nitrate (N4) + 2 mM ammonium (A2), 4 mM ammonium (A4), or 25 μg/mL paromomycin. Phenotypes of the different strains used are shown in the medium indicated.
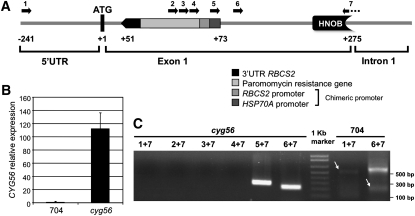
Insertional mutagenesis by marker DNA in Chlamydomonas causes results ranging from clean insertions to insertions involving deletions of several kilobases (Galvan et al., 2007). The cloning of the genomic regions flanking both sides of the insertion in mutant cyg56 was performed, and it was shown that the DNA marker had integrated in chromosome 16 (JGI v4.0). As indicated in Figure 3A, the insertion caused a 21-bp deletion at the first exon (position +51 after the translation start site) and disrupted a putative GC protein sequence. The ATG shown in this figure has the optimal nucleotide context for translation initiation in Chlamydomonas (Silflow, 1998; Kozak et al., 2002) and was presumed to be position +1. As a consequence of an rbcS2 promoter insertion, overexpression of the CYG56 transcript was detected at levels of ∼100 times those in the wild type (Figure 3B). This overexpression seems to result from the two enhancer elements of HSP70A promoter, a component of the transformation vector located at the vicinity of the RBCS2 promoter inserted in mutant cyg56, as reported by von Gromoff et al. (2006).
Figure 3.
Analysis of CYG56 Expression Following Insertion of the Paromomycin Resistance Marker in the AI Mutant cyg56.
The marker DNA is inserted near the 5′ terminal end of the CYG56 coding sequence.
(A) The vector pSI104 is inserted between positions 560341 and 560363 of scaffold 45 (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html) of the Chlamydomonas genome (corresponding to +51 and +72 of the coding region), just downstream of the initiation codon of CYG56 (annotated as +1) in the figure. Note that CYG56 is on the antisense strand of scaffold 45 in the Chlamydomonas genome. The parts of pSI104 marker are indicated.
(B) Expression of CYG56 was monitored by real-time PCR in the wild-type 704 and mutant cyg56. The data were obtained from a 3-h induction in 4 mM NO3− medium. Error bars represent the sd of three independent biological replicates.
(C) RT-PCR was performed in cyg56 and wild-type 704 using different combinations of primers whose locations are represented in (A).
RT-PCR amplifications (Figure 3C) using primers specific to the positions indicated in the scheme (Figure 3A) were performed to determine the 5′ end organization of the transcript expressed in mutant cyg56, and the amplified bands were sequenced. The 5′ end of the overexpressed CYG56 transcript is deleted in the mutant in the region containing the ATG codon and corresponds to a chimeric mRNA containing at least 40 bp of the insertional marker DNA fused, based on evidence that RT-PCR amplifications did not generate bands with primers 1, 2, 3, 4, and 7. Clear bands are amplified with primers 5+7 and 6+7 due to the overexpression of the transcript. Efficiency of the RT-PCR in the wild type was small because of the low expression level of CYG56 (Figures 3B and 3C), and this resulted in the appearance of nonspecific amplifications of high molecular weight bands when using primers 6 and 7.
Thus, these data indicate that a new transcription start site has been generated at the 3′ end of the inserted sequence in mutant cyg56 so that a partially deleted CYG56 transcript is expressed. The analysis of the insertion site also showed that sequence elements essential for translation are absent in the CYG56 chimeric transcript, ruling out the possibility that a functional CYG56 protein could be translated in the mutant. If the overexpressed CYG56 transcript was functional, the AI phenotype would have been obtained in diploids. This result of nonfunctional CYG56 was further confirmed by the data presented hereafter in this work. In summary, the linkage and complementation tests suggest a scenario in which lack of CYG56 functionality is responsible for the AI phenotype (i.e., NIA1 expression in the presence of ammonium).
CYG56 Encodes a Soluble GC Regulated by Nitrogen
A full-length CYG56 cDNA of 6111 bp was isolated and sequenced (EU841916). The sequence reveals an organization of 23 exons and 22 introns (see Supplemental Figure 1A online) that shows important differences with the coding region annotated in the genome (EDO98900; Merchant et al., 2007). Exons vary in size from 7 to 1950 bp, with most of the small exons comprising the 5′ half of the transcript. The cDNA has a long 3′ untranslated region with the typical TGTAA polyadenylation sequence that is common to other Chlamydomonas genes (Merchant et al., 2007).
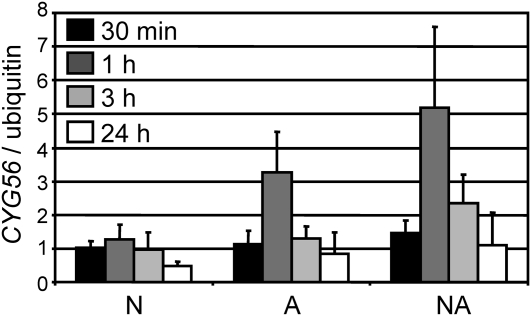
Transcriptional regulation of CYG56 by specific nitrogen sources was analyzed (Figure 4). CYG56 transcript levels increased in response to ammonium compared with nitrate as the sole nitrogen source. Interestingly, CYG56 showed a peak of expression at 1 h when cells were transferred to ammonium or nitrate plus ammonium. This weak but consistent activation of CYG56 expression is in agreement with a role for CYG56 in ammonium repression of NIA1.
Figure 4.
CYG56 Expression Is Activated by Ammonium in Wild-Type 704.
Wild-type 704 was grown in minimum medium containing 8 mM NH4Cl. After 3 d of growth, cells were induced in media containing 4 mM NO3−8 mM NH4+, and 4 mM NO3− + 8 mM NH4+. Samples were taken at 30 min and at 1, 3, and 24 h (black, dark-gray, light-gray, and white bars, respectively). RNA was extracted and CYG56 transcript levels were monitored by real-time PCR and normalized using the level of the transcript encoding ubiquitin. The error bars represent the sd of three biological replicates.
CYG56 encodes a protein (1721 amino acids) containing structural elements that are typical of animal NO-dependent GCs: HNOB (47 to 214), HNOBA (316 to 682), and GC catalytic domains (residues 692 to 822) (see Supplemental Figure 1B online). The HNOB domain is predicted to function as a heme-dependent sensor for gaseous ligands as NO and to transduce diverse downstream signals (Pellicena et al., 2004). HNOBA domain was recently shown to be involved in the dimerization of soluble guanylyl cyclase (Ma et al., 2008). This structure of globular N terminus extensions bearing HNOB and HNOBA domains before that of the catalytic GC seems to be an ancient conserved structure shared by animal soluble guanylyl cyclases and bacterial signaling proteins (Iyer et al., 2003; Nioche et al., 2004). In the deduced CYG56 sequence, a long C terminus of ∼900 amino acid residues is also present that is unusual in animal GCs and contains no characteristic domains. His binding heme and the invariable sequence motif YxSxR (Pellicena et al., 2004) are present in the HNOB domain (see Supplemental Figure 2A online). Some conserved Cys residues in HNOB and HNOBA domains that might be related to a strong activation of GC activity in animal GC (Fernhoff et al., 2009) are absent in Chlamydomonas GC (see Supplemental Figures 2A and 3 online).
The Chlamydomonas genome database contains 57 CYG genes, but only deduced proteins CYG11, CYG12, CYG15, CYG38, and CYG57 share the same domains structure with soluble CYG56 and GCs from animals (Nioche et al., 2004; Merchant et al., 2007), with an important degree of conservation (see Supplemental Figure 4 online). Scores of sequence identity obtained from comparisons of CYG56 with complete reported sequences range from 46% (with CYG57) and 24% (with CYG12) for Chlamydomonas proteins to 26% and 19% with proteins from animals. Interestingly, CYG11, CYG12, and CYG15 are linked within 50 kb at chromosome 7.
Both the full-length CYG56 protein and an N-terminal fragment of 957 amino acid residues were expressed in Escherichia coli, purified, and assayed for GC activity. The truncated protein was highly active and had a significant GC specific activity of 511 nmol·min−1·mg protein−1 in an assay with Mn2+ and 255 nmol·min−1·mg protein−1 using the physiological cation Mg2+ (see Supplemental Figure 5 online). However, it did not show any additional activation by the NO donors SNAP or DEA-NONOate described below. The full-length protein was partially purified and also showed a GC activity with a much lower activity assayed with Mg2+ relative to Mn2+ (see Supplemental Figure 5 online), and regulation by NO was not observed (data not shown).
The GC inhibitors ODQ, which is a selective inhibitor of NO-activated GC (Zhao et al. 2000), and LY-83,583 (LY) (Yang and Hatton, 1999) were used for in vitro assays with both the truncated and the full-length CYG56 proteins. ODQ showed a slight inhibition on GC cyclase activity assayed with Mg2+, but not with Mn2+, whereas LY caused a 60 and 45% inhibition in the activity of truncated and full-length CYG56 proteins, respectively, in assays with Mg2+, and much less with Mn2+ (see Supplemental Figure 6 online). The existence of long loops of amino acids disrupting the HNOBA domain in CYG56 might represent a difficulty for a proper folding in E. coli, resulting in a protein with a folded GC catalytic domain that is insensitive to signals from heme domains, and thus to possible activation by NO or inhibition by ODQ. Alternatively, another protein subunit could be required for assembly of an adequate functional complex that has specific NO control of GC activity. The lack of conserved Cys residues able to be nitrosylated (see Supplemental Figures 2 and 3 online; Fernhoff et al., 2009) might also result in weaker activation than in animal GCs. The presence of highly conserved HNOB and HNOBA and the series of experiments detailed below strongly suggest that CYG56 should be activated by NO in vivo.
NO and cGMP Repress NIA1 Transcription in the Chlamydomonas Wild-Type Strain 704
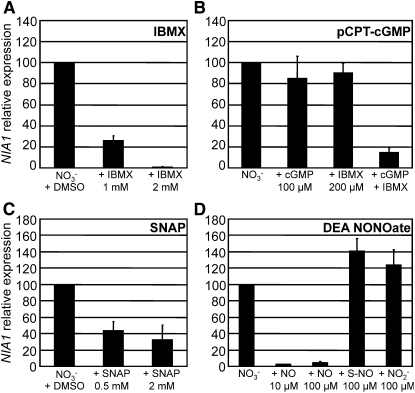
To assess whether NO and cGMP indeed repress NIA1 transcription, a set of pharmacological products that increase intracellular concentrations of these molecules were applied to Chlamydomonas cell cultures. The phosphodiesterase inhibitor isobutylmethylxanthine (IBMX), the cGMP analog pCPT-cGMP, and NO donors SNAP and DEA NONOate were used, and their effects on NIA1 expression were measured by real-time PCR. In the wild-type 704, IBMX inhibited NIA1 transcription in a dose-dependent manner. Application of 1 mM IBMX led to a 75% decrease of NIA1 transcript levels compared with the untreated control, and 2 mM IBMX led to almost full inhibition of gene expression (Figure 5A). The cGMP analog pCPT-cGMP at 100 μM had a minor effect; however, when combined with 200 μM IBMX, a concentration at which IBMX alone did not show any significant effect, this cGMP analog caused a strong inhibition of NIA1 expression (Figure 5B). The NO donor SNAP also reduced wild-type NIA1 transcript levels to 32% when used at 2 mM (Figure 5C). DEA NONOate, a more potent NO donor, caused a full inhibition of NIA1 expression at concentrations as low as 10 μM, whereas 100 μM of the analog SulphoNONOate that does not produce NO had no effect. Nitrite derived from oxidation of NO (Ignarro et al., 1993) does not seem to be responsible for the observed effects of DEA NONOate since application of 100 μM nitrite had no effect (Figure 5D). Taken together, these results suggest that cGMP and NO are able to repress NIA1 transcription.
Figure 5.
IBMX, pCPT-cGMP, SNAP, and DEA NONOate Repress NIA1 Transcription in the Wild-Type 704.
The effect of IBMX (A), pCPT-cGMP (8-(4-chlorophenylthio)guanosine-3′,5′-cyclic monophosphate) (B), SNAP (S-nitroso-N-acetylpenicillamine) (C), and diethylamine NONOate (DEA NONOate) (D) on NIA1 transcription was determined by real-time PCR. The effect of DEA NONOate (NO) was compared with those of chemical control SulphoNONOate (S-NO) and nitrite at the indicated concentrations. The error bars represent the sd of at least three replicates, with each replicate corresponding to a different PCR run. As IBMX and SNAP are soluble in DMSO, DMSO was added in the untreated control. cGMP represents pCPT-cGMP.
Other Ammonium-Repressible Nitrogen Assimilation Genes Respond to NO and cGMP
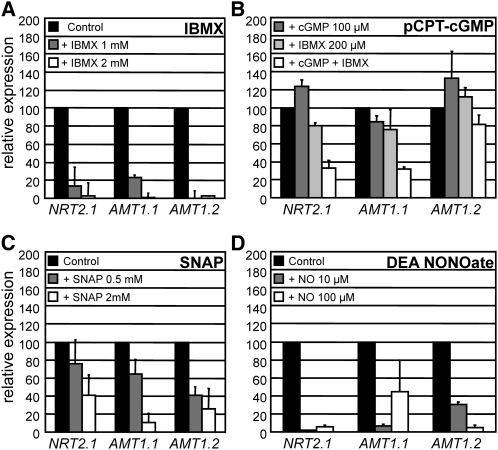
NIA1 and NRT2.1, which encodes a component of a high affinity nitrate transporter, have in common that they are repressed by ammonium and are positively regulated by the nitrate-specific regulatory protein NIT2 (Llamas et al., 2002; Rexach et al., 2002). AMT1.1 and AMT1.2 encode ammonium transporters, and their expression levels are also significantly reduced in ammonium compared with nitrate. As for NIA1 and NRT2.1, AMT1.1 expression was shown to be regulated by NIT2 (González-Ballester et al., 2004). To assess whether the effect of NO and cGMP was specific for NIA1 or whether they affected nitrate regulated genes in a more general way, we tested the effect of IBMX, pCPT-cGMP, and NO donors on the expression of NRT2.1, AMT1.1, and AMT1.2. As for NIA1, IBMX, pCPT-cGMP, SNAP, and DEA NONOate caused repression of NRT2.1, AMT1.1, and AMT1.2 in a dose-dependent manner (Figures 6A to 6D). Again, 100 μM pCPT-cGMP had a negative effect on the expression of those genes when combined with 200 μM IBMX. Another cGMP analog, 8-bromo (8Br)-cGMP, had similar negative effects on expression of NIA1, NRT2.1, and AMT1.1 as the Cl-derivative (i.e., minor when applied alone and significant in combination with 200 μM IBMX). However, 8-Br-cGMP had no effect on expression of AMT1.2 (see Supplemental Figure 7 online). The minor effects of cGMP analogs on nitrate assimilation gene expression can be explained by hydrolysis of these compounds by phosphodiesterases, that when inhibited resulted in the observed effects (Boss, 1989). This also shows that repression of NIA1 by IBMX is most likely due to the upregulation of intracellular cGMP rather than cAMP.
Figure 6.
Effect of IBMX, pCPT-cGMP, SNAP, and DEA NONOate on Expression of Ammonium-Repressed Genes NRT2.1, AMT1.1, and AMT1.2 in Wild-Type Cells.
The effect of IBMX (A), pCPT-cGMP (B), SNAP (C), and DEA NONOate (D) on the expression of NRT2.1 and AMT1.1 and AMT1.2 was determined by real-time PCR. Conditions of the experiment were the same as in Figure 5 (see Methods). Black bars represent controls (set to 100%) without the added chemical; gray and white bars represent the concentrations of each chemical as indicated in each panel. The error bars represent the sd of three biological replicates.
DEA NONOate also proved to be a more potent NO donor than SNAP as concentrations as low as 10 and 100 μM showed a strong negative effect on the expression of those three genes (Figures 6C and 6D). Although the expression of the housekeeping gene ubiquitin ligase (see Methods) remained unaltered by these treatments, we tested the expression of actin as an additional control to check whether transcription was affected by DEA NONOate. The expression levels of actin remained unaltered after the treatments (data not shown). Taken together, these data suggest that cGMP and NO mimic the effects of ammonium on expression of these genes.
The GC Inhibitors, LY and ODQ, and the NO Scavenger cPTIO Release Repression by Ammonium and NO, Respectively, in the Wild-Type Strain
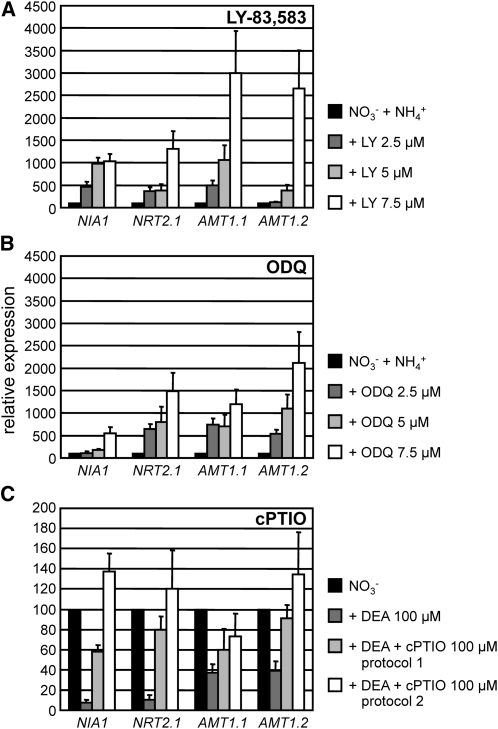
To support better that NO-dependent GC has a role on ammonium repression, we reproduced a deficiency of GC activity in the wild-type strain using the GC inhibitors LY and ODQ. The inhibitors were applied in medium containing NO3−NH4+ (in which NIA1, NRT2.1, AMT1.1, and AMT1.2 transcript levels were low). As expected, expression of these four genes was derepressed by LY treatments in a dose-dependent manner (Figure 7A). Similarly, ODQ also caused derepression of the four genes in ammonium containing medium (Figure 7B). As shown in Figures 5 and 6, NO supplied by DEA-NONOate promoted a strong inhibition of nitrate induction for these genes. The effect of the specific NO scavenger cPTIO (Planchet and Kaiser, 2006) was determined in two different ways: first, by preincubating the cells with cPTIO during 15 min before adding DEA (protocol 1, Figure 7C) and by preincubating DEA and cPTIO together before applying them to the cells (protocol 2, Figure 7C). Both protocols were efficient in inhibiting the effect of DEA.
Figure 7.
Treatment of Wild-Type Cells with GC Inhibitors LY and ODQ Leads to Partial Release of NIA1, NRT2.1, AMT1.1, and AMT1.2 Expression in Media Containing Nitrate and Ammonium: Effect of the NO Scavenger cPTIO.
The effect of LY83,583 (6-anilino-5,8-quinolinedione) and ODQ (1H-[1,2,4]oxadiazolo-[4,3-a]quinoxalin-1-one) on the expression of the indicated genes was determined by real-time PCR. Wild-type 704 was grown in minimum medium containing 8 mM NH4Cl. After 3 d of growth, cells were induced during 1 h in minimum medium containing 4 mM KNO3 + 1mM NH4Cl supplied LY (A) or ODQ (B) at the concentrations indicated. Alternatively (C), expression of these genes was determined in cells induced during 1 h in minimum medium containing 100 μM KNO3 in the absence or presence of NONOate 100 μM (used as controls). The effect of 100 μM cPTIO was analyzed either after preincubating the cells 15 min with cPTIO previously to NONOate addition (protocol 1) or when added simultaneously with DEA NONOate at time 0 (protocol 2). The error bars represent the sd of three biological replicates.
These data so far suggest that (1) repression of nitrogen assimilation gene expression by NH4+ at least partially requires an NO-dependent GC activity, and (2) NH4+, NO, GC, and cGMP are components of a common regulation pathway that represses NIA1, NRT2.1 AMT1.1, and AMT1.2.
Ammonium Enhances NO and cGMP Levels in Chlamydomonas Cells
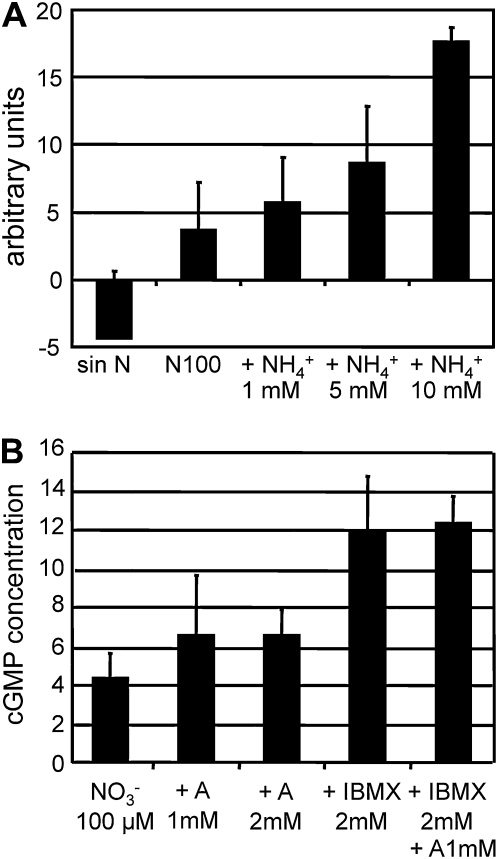
If NO is implicated in the repression of NIA1 by ammonium, then we predicted that ammonium would increase NO intracellular levels. To assess this, intracellular NO was quantified by fluorescence using the DAF-FM DA (4-amino-5-methylamino-2'7'-difluorofluorescein diacetate) dye (Kojima et al., 1998; Planchet and Kaiser, 2006). As shown in Supplemental Figure 6 online, DAF-FM DA only emitted fluorescence after entering the cells and fluorescence was directly proportional to the amount of applied DEA NONOate. NO production was measured after different nitrogen treatments of the cells (Figure 8). Cells transferred to media containing nitrate plus different amounts of ammonium showed that stimulation of NO production depended on the ammonium concentration (Figure 8A). NO production was higher in cells coming from a preculture in N-free medium for 24 h (see Supplemental Figure 8B online) than in cells coming from the standard ammonium medium (Figure 8A). These cells from N-free medium did respond slightly to increasing nitrate concentrations though much less than to ammonium (see Supplemental Figure 8B online). However, in media containing both 5 mM ammonium and increasing concentrations of nitrate, the observed NO production mostly resulted from ammonium with little appreciable effect of the high concentrations of nitrate (see Supplemental Figure 8C online).
Figure 8.
Ammonium Increases Intracellular Levels of NO and cGMP.
Fluorescence due to intracellular NO levels after incubation with the indicated nitrogen sources was determined using DAF-FM DA and is expressed as arbitrary units.
(A) Cells were grown in 8 mM ammonium medium, washed, and transferred to N-free medium and media containing 100 μM nitrate plus the indicated concentration of ammonium for 1 h, and then fluorescence was determined.
(B) cGMP concentration was determined from cells grown in 8 mM ammonium (A) medium, washed, and transferred for 1 h to the indicated medium. Amounts of cGMP are expressed as fmol/106 cells. The error bars represent the sd of three biological replicates.
Levels of cGMP in the cells were also determined in response to treatments with IBMX or ammonium. As shown in Figure 8B, cGMP levels in cells incubated in 100 μM nitrate increased slightly due to the presence of ammonium 1 or 2 mM. However, these levels increased significantly after 1 h treatment with 2 mM IBMX. There was an insignificant effect of added ammonium, probably due to the fact that, from the high number of GCs in Chlamydomonas (Merchant et al., 2007), only a few of the GCs mediate ammonium effects. These results reinforce that ammonium effects are mediated by NO and cGMP.
Ammonium-Repressible Nitrogen Assimilation Genes Respond to Ca2+ and to L-NAME, an Inhibitor of NO Synthesis
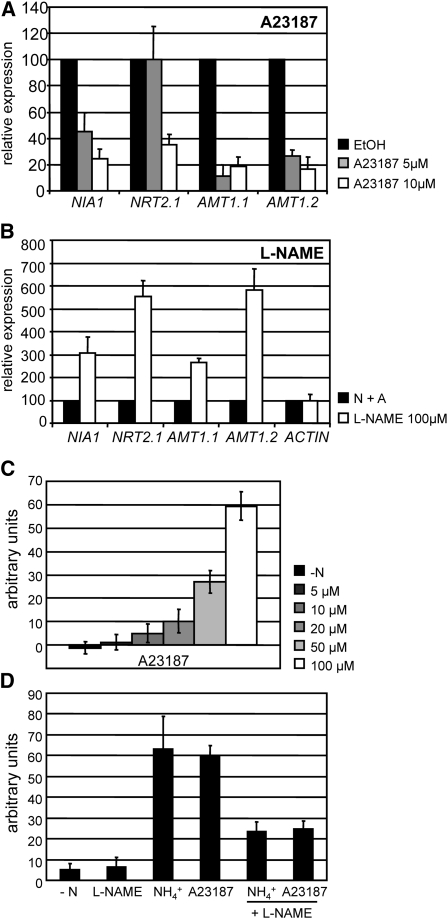
In plants, synergistic effects of Ca2+, NO, and cGMP in various physiological processes, such as response to pathogen infection and development of cell polarity, have been reported (Sokolovski et al., 2005; Lanteri et al., 2006; Ali et al., 2007; Salmi et al., 2007). Implication of these second messengers in the same regulation pathways is often associated with Ca2+ being an activator of a possible nitric oxide synthase (NOS). We tested whether Ca2+ was also implicated in the regulation of NIA1 and other ammonium-repressible nitrogen assimilation genes by measuring the effects of the calcium ionophore A23187 on their transcript levels. Addition of A23187 reduced NIA1, NRT2.1, AMT1.1, and AMT1.2 expression in a dose-dependent manner, as 5 and 10 μM of A23187 resulted in a decrease in their transcript levels relative to untreated controls (Figure 9A). As for SNAP and IBMX, AMT1.1 and AMT1.2 were more sensitive to A23187. Though side effects caused by this calcium channel inhibitor cannot be ruled out, these data suggest that increases in intracellular Ca2+ levels also repress nitrogen assimilation gene transcription. On the other hand, L-NAME, an inhibitor of NO synthesis (Radomski et al., 1990), caused a partial release of the negative effects of ammonium on NIA1, NRT2.1, AMT1.1, and AMT1.2 expression (Figure 9B). Expression of actin, used as a control, was not affected by the presence of this inhibitor.
Figure 9.
Effect of A23187 and L-NAME on Ammonium Repression of Gene Expression and on the Levels of NO.
The effect of A23187 (A) and L-NAME (B) on the expression of NIA1, NRT2.1, AMT1.1, and AMT1.2 was determined by real-time PCR. Wild-type 704 was grown in minimum medium containing 8 mM NH4Cl. After 3 d of growth, cells were induced during 1 h in minimum medium containing 100 μM KNO3 in (A) or 100 μM KNO3 + 1 mM NH4Cl in (B) and treated with the chemicals at the concentrations indicated. The effect of A23187 (C) and L-NAME (D) on the fluorescence levels due to intracellular NO determined using DAF-FM DA and expressed as arbitrary units. Cells preincubated for 24 h in N-free medium were transferred to the indicated N-free medium containing the indicated concentration of A23187 (C) or 5 mM ammonium, 100 μM A23187, and 100 μM L-NAME, as indicated (D). The error bars represent the sd of three biological replicates.
Interestingly, the Ca2+ ionophore promoted NO production in a dose-dependent manner (Figure 9C). L-NAME inhibited efficiently NO production resulting from either the treatment with ammonium or the calcium ionophore (Figure 9D). These results suggest that Ca2+ and a putative NOS could be involved in the negative regulation of ammonium regulated genes mediated by NO.
Mutant cyg56 Is Altered in a Signaling Pathway That Involves Ammonium, NO, cGMP, and Ca2+
Comparative studies between the wild type and mutant cyg56 using effectors upstream and downstream of the GC enzyme in the signaling pathway were useful to better understand this pathway. Results of effectors are presented as matrices of gene expression where transcript levels are normalized to the untreated control and the level of repression is represented by a scale of shading (Figure 10; see Supplemental Table 1 online). Consistently, results showed that SNAP, an NO donor, did not affect expression of NIA1 and NRT2.1, in mutant cyg56 in contrast with the wild type. However, IBMX repressed NIA1 and NRT2.1 expression to a similar extent in both the mutant and the wild-type 704 (Figures 10A and 10B).
Figure 10.
Expression of NIA1 and NRT2.1 Is Partially or Completely Derepressed in Mutant cyg56 Cells When Treated with SNAP, A23187, and SNP but Not with IBMX and DEA NONOate.
Relative transcript levels of NIA1 (A) and NRT2.1 (B) were quantified by real-time PCR in strains 704 and cyg56 when treated with the indicated pharmacological products. Concentrations were 1 mM SNP and FeCN, 10 μM DEA NONOate, 2 mM IBMX, 2 mM SNAP, and 10 μM A23187. Data are summarized in matrices of gene expression in which relative transcript levels are represented by a gray code. In both tables, each column corresponds to a strain and each row corresponds to the effect of a pharmacological product. Results from the treated samples were normalized to the untreated control for each PCR run. Conditions of the experiment were the same than in Figure 5. Raw data are presented in Supplemental Table 1 online.
To strengthen these results, this experiment was performed with the NO donor sodium nitroprusside (SNP). The effects of SNP were compared with ferricyanide (FeCN) as a chemical control since both components contain cyanide in their molecular structure (five molecules of cyanide for SNP and six for FeCN). The difference between these two is the presence of a nitrous group responsible for the NO-generating property of SNP (Oh and McCaslin, 1995; Bethke et al., 2005). SNP strongly repressed NIA1 and NRT2.1 transcription in the wild type, but this effect was weaker in mutant cyg56 (Figures 10A and 10B). Surprisingly, FeCN also repressed the expression of these genes, which suggested that cyanide acted negatively on NIA1 and NRT2.1, but this effect was similar in 704 and cyg56. These results argued in favor of mutant cyg56 being altered in a pathway specific for NO-mediated NIA1 and NRT2.1 repression that is independent of cyanide.
Unexpectedly, the strong NO donor DEA NONOate caused repression of NIA1 and NRT2.1 in both 704 and cyg56 as derepression of the genes in the mutant was very mild (see Supplemental Table 1 online). The repression of NIA1 by DEA NONOate in cyg56 might be directly related to the higher potency of this NO donor, a more efficient release of intracellular NO possibly leading to activation of other GCs that are also implicated in NIA1 repression. This idea is supported by the presence of >50 predicted GC domains in the genome of Chlamydomonas (Merchant et al., 2007) from which at least six are NO-dependent type GC as shown above.
Finally, calcium ionophore A23187 had a slight effect on expression of NIA1 and NRT2.1 transcripts in mutant cyg56 in contrast with the wild type (Figures 10A and 10B; see Supplemental Table 1 online). Taken together, these results strongly suggest that NH4+, NO, cGMP, and Ca2+ act in a common regulation pathway that represses NIA1 and NRT2.1 transcription and that this pathway is altered in mutant cyg56. Furthermore, opposing effects of A23187and NO donors with IBMX in mutant cyg56 suggest that Ca2+ and NO act upstream of cGMP in this pathway.
NO Represses NIA1 and NIA2 Transcription in Arabidopsis
Genes and mechanisms of the nitrate assimilation pathway are well conserved between Chlamydomonas and higher plants; thus, cGMP and NO could also be signal molecules for repression of nitrate assimilation genes in Arabidopsis. First, published microarray data (Maathuis, 2006) revealed that application of 10 μM 8-Br-cGMP to Arabidopsis roots reduced NIA2 expression to 35% relative to the untreated control 5 h after exposure. NRT2.1 and NRT2.2 mRNA levels were also reduced to 46 and 33%, respectively, in the same conditions. Moreover, AMT1 was even more sensitive as expression levels of this gene dropped to 27% 2 h only after exposure to 8-BrcGMP. These data suggested that cGMP represses nitrogen assimilation genes in Arabidopsis.
Here, we analyzed the effect of an NO donor on the expression of genes encoding NRs in Arabidopsis by applying DEA NONOate to 10-d-old seedlings. As shown in Figure 11, both NIA1 and NIA2 are repressed in a time-dependent manner by NO in comparison to control plants maintained in the medium with no additions. The chemical control SulfoNONOate did not affect expression of either of the two genes. These results show that NIA1 and NIA2 gene expression is sensitive to NO and suggest that, in Arabidopsis, mechanisms similar to those found in Chlamydomonas involving NO on the negative control of nitrate assimilation genes might be operative.
Figure 11.
NO Represses NIA1 and NIA2 from Arabidopsis.
The effect of DEA NONOate and SulphoNONOate on At NIA1 (A) and At NIA2 (B) expression was tested by real-time PCR in 10-d-old Arabidopsis seedlings. DEA NONOate and SulphoNONOate (100 μM) was applied directly on the seedlings as described in Methods. Plant material was harvested before treatment (black bars) and after 30 min (gray bars) and 1 h (white bars) as indicated. The error bars represent the sd of three biological replicates.
DISCUSSION
Regulation of nitrate assimilation involves the integration of positive signals (i.e., nitrate) and negative signals (i.e., ammonium and ammonium derivatives), which result in appropriate use of the nitrogen source. In this work, we analyzed negative signaling by ammonium on NIA1 expression using a Chlamydomonas wild-type strain and a mutant defective in a putative GC. The results allow us to conclude several points as described in the following sections.
Ammonium Repression of NIA1 Appears to Be a Complex Process
Ammonium repression of NIA1 is shared by the other genes for nitrate assimilation in Chlamydomonas (Fernandez and Galvan, 2007), and this negative regulation by ammonium assimilation derivatives is also observed in other plant systems (Stitt, 1999; Crawford and Forde, 2002; Glass, 2003; Fan et al., 2006). That this is a complex process can be deduced not only from data in the bibliography but also from results generated in Chlamydomonas. First, the screening of a Chlamydomonas insertional mutant library produced a set of mutants that showed different AI phenotype intensities and that were defective in different genes for ammonium sensing (González-Ballester et al., 2005a). Second, the AI phenotype depends on the concentration of ammonium in the medium, so that more than one mechanism might mediate ammonium sensing at diverse concentrations as previously proposed. Finally, of all the AI mutants isolated from that screen or from other previously reported screens (Prieto et al., 1996; Zhang and Lefebvre 1997; Perez-Alegre et al., 2005), none was fully insensitive to ammonium. One of the goals of this work was the analysis of one of those mutant strains that was affected in a putative GC, since that might reveal a relationship between regulation of nitrate assimilation and cGMP.
An NO-Dependent Guanlylate Cyclase Is Responsible for the AI Phenotype of Mutant cyg56
Insertion of a marker gene in the AI mutant cyg56 caused a critical deletion at the 5′ end of the CYG56 gene, fusing it to the sequence of the maker in a chimeric transcript that is overexpressed. These modifications are compatible with a loss of function of CYG56 gene. This is also supported by the complementation of the mutant phenotype in diploids and the reproduction of an AI phenotype by the GC inhibitors LY and ODQ in the wild-type strain.
CYG56 expression was upregulated by ammonium plus nitrate medium, and the maximum level of expression was reached in 1 h, which indicates that upregulation of CYG56 by nitrate plus ammonium could be transient. This regulation suggests a role of CYG56 when both sources of nitrogen are present. In other terms, in ammonium plus nitrate medium, CYG56 signaling on NIA1 makes sense, since NIA1 negative control by ammonium can only take place after expression has been switched on in nitrate (Camargo et al., 2007).
CYG56 is a NO-dependent GC containing the three domains typical of this kind of GCs that are present in animals and absent in plants (Nioche et al., 2004). The enzyme activity of CYG56 as a GC was demonstrated using both the full-length protein and the N terminus protein fragment of 957 residues containing the GC catalytic domain and the domains for NO binding. However, the expressed proteins already had a high GC activity so that additional NO activation could not be shown, possibly due to an inappropriate folding of this protein in the E. coli host that would release the GC domain from the control by NO. Additionally, another subunit might be required to make the GC domain further activated by NO as reported in other animal GCs (Koglin et al., 2001). Interestingly, CYG56 lacks three Cys residues conserved in other GCs (see Supplemental Figures 2 and 3 online). Nitrosylation of Cys residues has been implicated in the strong activation by NO of animal GC versus the moderate activation caused by NO binding to the heme group (Fernhoff et al., 2009). The fact that at least six GCs from the Chlamydomonas genome (Merchant et al., 2007) have the same domain structure as CYG56 could suggest a moderate activation of GCs, and this would be necessary for a more precise and fine regulation of gene expression. It is also interesting that of these six members of the gene family encoding NO binding GC, three (CYG11, CYG12, and CYG15) are clustered in the same chromosome 7 (CYG56 is in chromosome 16).
Thus, a signal transduction protein typical of animals has been selected in Chlamydomonas for regulating a metabolic pathway, nitrate assimilation, typical of plants and bacteria but not of animals. Guanylyl cyclases that are not well conserved with animal GCs have been described in higher plants (Ludidi and Gehring, 2003; Yuan et al., 2008) and proteins having a GC domain with a modular structure such as the brassinosteroid receptor described (Kwezi et al., 2007). However, NO and cGMP appear to regulate numbers of physiological processes in plants. A possibility is that proteins involved in sensing belong to diverse protein families or have particular combinations of domains to deal with similar regulatory circuits or strategies in the different organisms.
A Regulation Model for NIA1 Transcription Involves Ammonium, Calcium, NO, and cGMP
This work provides pharmacological and genetic evidences that ammonium, calcium, NO, and cGMP act by a similar pathway to repress NIA1 and NRT2.1 and that this process requires GC activity. The facts supporting that NO is a component of the ammonium-CYG56-cGMP negative signaling pathway are (1) NO represses expression of NIA1, NRT2.1, AMT1.1, and AMT1.2. This was shown with the three NO donors SNAP, SNP, and DEA-NONOate. No effect was seen with the chemical control SulfoNONOate. In addition, DEA-NONOate had no effect after preincubation with cPTIO. (2) CYG56 has a conserved NO binding domain. (3) Ammonium increases NO intracellular levels. (4) ODQ blocks the repression of NIA1 by ammonium; and, most importantly, (5) the effect of SNAP and SNP on the expression of NIA1 and NRT2.1 does not take place in the cyg56 mutant.
The facts supporting that cGMP is a component of the ammonium-CYG56-cGMP negative signaling pathway are (1) IBMX and ammonium increase intracellular levels of cGMP. (2) cGMP analogs (pCPT-cGMP and 8-Br-cGMP) cause repression of NIA1 and NRT2.1. (3) Inhibition of the NO-dependent GC by LY or ODQ releases cells from ammonium repression of NIA1, NRT2.1, AMT1.1, and AMT1.2, reproducing the phenotype of mutant cyg56. Published microarray data from Arabidopsis suggested that this regulation pathway could be conserved in higher plants as application of 8-Br-cGMP repressed NIA2, NRT2.1, NRT2.2, and AMT1 (Maathuis, 2006).
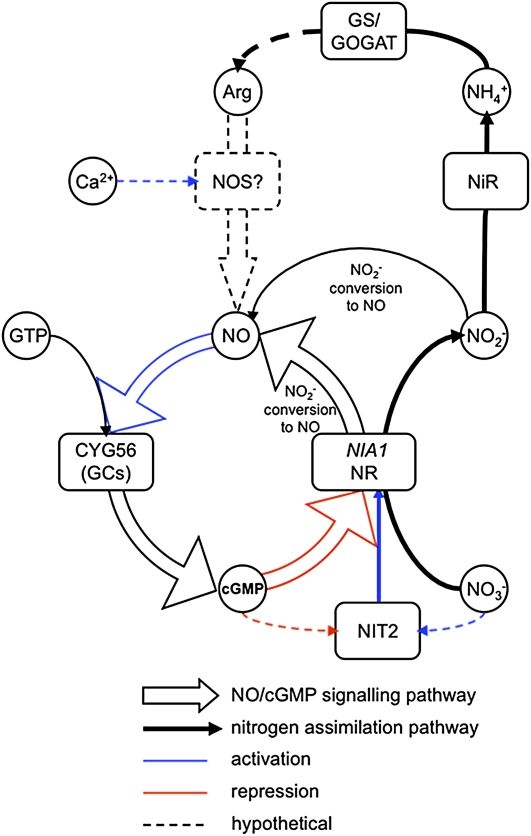
Based on results presented here and on data from the literature, we propose a regulation model for the repression of NIA1 that integrates ammonium, calcium, NO, and cGMP (Figure 12). In this model, CYG56 has a central role. Ammonium is generated by sequential reduction of nitrate and nitrite through NR and NiR activities, respectively, or can be transported from the extracellular medium (Galvan et al., 2006). Ammonium is integrated into the structure of amino acids by the GS/GOGAT cycle, leading in turn to the synthesis of Arg (Crawford, 1995; Slocum, 2005). Hypothetically, Arg could be converted to NO and citrulline by a putative NOS activity, Ca2+ being involved in this process as an activator of the enzyme. NO would then activate one or several GCs that produce cGMP from GTP, and cGMP would in turn repress NIA1 transcription. A similar process would be operative for NRT2.1. The positioning of NO and Ca2+ upstream of CYG56 is mainly supported by the A23187, SNAP, and SNP-mediated repression of NIA1 being partially compensated in the cyg56 mutant. In support of this model is the dose-dependent NO production promoted by Ca2+ ionophore and its inhibition by L-NAME, in a similar manner of the NO production promoted by ammonium.
Figure 12.
Proposed Scheme for NIA1 Negative Regulation Involving Ammonium, Arg, Ca2+, NO, cGMP, and Nitrite.
The regulation scheme involves the following steps: (1) Nitrate is reduced to nitrite and nitrite to ammonium by sequential action of NR and nitrite reductase (NiR). Ammonium can also be transported inside the cell from the extracellular medium. (2) The GS/GOGAT cycle integrates ammonium to the metabolism leading to the synthesis of amino acids. (3) In plant systems, Arg is converted to NO through Ca2+ dependent of a putative NOS activity, but the corresponding NOS gene is so far unknown, which is why this step is presented as hypothetical in the model. (4) NO activates CYG56 and maybe other soluble GCs that catalyze the formation of cGMP. (5) cGMP represses NIA1 transcription, maybe by interfering with NIT2 functionality. (6) NO can be generated by enzyme (NR activity or mitochondrial electron transport) or chemical conversion of nitrite.
The mechanism by which cGMP could regulate NIA1 transcription is unclear. Putative cGMP-dependent protein kinases and their targets are not known. The presence of a GAF domain on the sequence of the NIA1 positive regulator NIT2 suggests a possible role for cGMP in mediating NIT2 functionality (Camargo et al., 2007). This idea is supported by NIT2 being implicated in the regulation of NIA1, NRT2.1, and AMT1.1. As NO and cGMP repress the expression of all three genes, a possibility is that cGMP binds to NIT2, leading to its inactivation. The GAF domain was shown not to participate in DNA binding of NIT2 (Camargo et al., 2007), but it could be implicated in conformational changes of the protein. Interestingly, NO has also been shown to bind GAF domains of NorR protein (Busch et al., 2005). This signaling on NIT2 might occur at some other step.
An essential question that arises from the model and from our data is: By which mechanism NO is actually synthesized? Well-known sources of NO in plants and other organisms are Arg and nitrite. NOS activity dependent on Arg with citrulline formation has been observed in plant extracts, as its inhibition by Arg analogs (Cueto et al., 1996; Durner et al., 1998; Delledonne et al., 1998; Del Rio et al., 2004).
Arg is converted to NO through the activity of a possible NOS, a controversial enzyme that has not yet been cloned in plant systems (Guo et al., 2003; Zemojtel et al., 2006). The potential Arabidopsis NOS1 previously identified corresponds to a 561–amino acid protein having no sequence homology to the animal NOSs (Guo et al., 2003). At NOS1 has been renamed At NOA1 and shown to be a functional Arabidopsis member of the circularly permuted GTPase family (cGTPase) with no NO synthase activity (Moreau et al., 2008). In our experiments, evidence of NOS activity being implicated in the described model is given by the partial release of NIA1 repression by L-NAME, an inhibitor of NOS activity. Moreover, the repression of NIA1 by A23187 in the wild type but not in the mutant suggests the involvement of Ca2+ in NIA1 regulation upstream of CYG56. These results support the implication of NOS in the model as Ca2+ is an activator of NOS in animals, and NOS activity has been shown to be Ca2+ dependent in plants (Delledonne et al., 1998; Courtois et al., 2008). Nevertheless, this evidence remains insufficient to demonstrate that NOS is the source of NO in the ammonium-mediated NIA1 repression pathway as NOS activity has not yet been shown in Chlamydomonas and considering that the existence of this enzyme remains uncertain in plants. The presence of the enzyme in the model is therefore stated as hypothetical. Future experiments must address this question.
Nitrite can be converted to NO through either NR activity or the mitochondrial electron transport chain (Yamasaki and Sakihama, 2000; Bethke et al., 2004; Tischner et al., 2004; Planchet et al., 2005). Interestingly, nitrite was shown to repress transcription of NIA1 (Loqué et al., 2003), raising the question of whether nitrite-mediated NIA1 repression is due to nitrite conversion to NO. The particular conditions regulating NR activity for nitrite conversion to NO in planta are not known. Lamattina et al. (2003) suggested that changes in NR activity could be modulated by nitrite availability. When the plant is photosynthetically active, nitrite produced by NR is immediately taken up by the chloroplast and reduced to ammonium (Rexach et al., 2000). However, nitrite accumulation can be expected when photosynthetic activity is low, so that NR-mediated conversion of nitrite to NO could take place in these conditions (Lillo et al., 2003; Lea et al., 2004) and NO could then repress NIA1 transcription, the whole process forming a feedback loop. This negative feedback loop could be part of a mechanism for protection against nitrite toxicity, so that further accumulation of nitrite would be blocked.
Another important question that arises from our data is whether other GCs than CYG56 are implicated in the repression of NIA1. First, this idea is supported by the presence of >50 predicted GC catalytic domains in the genome of Chlamydomonas (Merchant et al., 2007). A report proposing the presence of a similar amount of GCs in the genome of Arabidopsis, although with limited sequence conservation (Kwezi et al., 2007), suggests a similar situation in higher plants. Second, the cyg56 AI phenotype was mild in comparison with that of other mutants isolated from the initial screen (González-Ballester et al., 2005a). In cyg56, other GCs could still be repressing NIA1 expression in response to ammonium, explaining why the phenotype was partial. Third, further evidence was given by the variety of responses obtained after treatment with different NO donors. SNAP was the less efficient NO donor tested, as the half-life of this compound is of several hours, whereas the half-life of DEA NONOate is of 17 min in controlled pH conditions. Consistently, application of DEA NONOate had drastic effects on NIA1 expression, whereas the effect of SNAP was weaker. Moreover, release of NIA1 repression in cyg56 after treatment with SNAP was almost total and contrasted with the weak derepression observed with DEA NONOate. A possibility is that the stronger NO donor activates more efficiently other GCs implicated in the repression of NIA1 that are not activated by SNAP. Taken together, this evidence suggests that various GCs being implicated in the ammonium-mediated repression of NIA1 is a possible scenario that should also be addressed by future experiments.
In conclusion, we provide genetic evidence on that a NO-dependent GC of animal type is involved in the negative control of the nitrate assimilation pathway in Chlamydomonas. The effect of treatments with pharmacological compounds on gene expression allowed us to propose that ammonium, Ca2+, NO, and cGMP act in a common pathway that represses NIA1 and other ammonium-repressed genes. We also show that ammonium enhances the intracellular production of NO and cGMP. Evidence for a similar process to occur in plants is provided. A mechanistic approach is needed to understand how cGMP represses the transcription of ammonium-repressed genes.
METHODS
Strains and Culture Conditions
The Chlamydomonas reinhardtii strain 704 (cw15 arg7+ NIA1:Ars mt+) (Loppes et al., 1999) is the parental type of the AI mutant strain 20.73, named here as cyg56 (mt+cw15 arg7+ NIA1:Ars rbsc2:AphVIII) and obtained by insertional mutagenesis as previously detailed (González-Ballester et al., 2005a). Mutant strains 305 (NIA1- mt-) and 203 (Nit2- mt+) have been characterized elsewhere (Fernandez and Matagne, 1984). Cells were cultured under continuous light at 23°C in liquid or solid minimum medium (Harris, 1989) containing different concentrations of potassium nitrate depending on the experiment and were bubbled with 5% (v/v) CO2-enriched air.
Arabidopsis thaliana Columbia-0 was grown on MS medium (Murashige and Skoog, 1962) in growth chambers providing controlled temperature (22°C) and a daylength regime of 16 h of light and 8 h dark per day. For the experiment shown in Figure 11, plants were grown in 12-well plates containing 2 mL of solid MS medium. Plant material harvested at different time points was grown in separate wells. Untreated, SulphoNONOate-treated and DEA NONOate-treated plants were grown in separate plates.
Genetic Analyses and Complementation Tests
Genetic crosses were performed using the random spore plating method (Harris, 1989). Segregants were analyzed for their ability to grow on medium containing nitrate by replica plating, for their resistance to 25 μg/mL paromomycin, caused by the inserted APHVIII gene from the marker, and for the AI phenotype as described (González-Ballester et al., 2005a). In vivo complementation was performed by isolating stable vegetative diploids according to the method of Ebersold (Fernández and Matagne, 1986; Harris, 1989).
ARS Activity
ARS activity was determined directly on the agar plates after removing cells from the agar surface with a razor blade and following the procedure previously reported (Ohresser et al., 1997). The reaction was stopped after 15 min.
NR Activity
NR activity was determined in vitro following the procedure previously described. Cells (100 μL) were lysed with 5 μL of 100% toluene (Fernandez and Cardenas, 1982). Benzyl viologen reduced with dithionite was the electron donor. The reaction was stopped by vortexing of the tubes, and nitrite, the product of NR activity, was determined as described by Snell and Snell (1949). Activity values were expressed as milliunits (mU), defined as the amount of enzyme catalyzing the transformation of 1 nmol of substrate per min, relative to the chlorophyll content, determined as described by Arnon (1949).
cGMP Determination and GC Expression and Activity
Both a full-length CYG56 protein and its N-terminal fragment of 957 amino acid residues were expressed in Escherichia coli using the vector pQE80 (Qiagen) that produces proteins labeled with a 6xhis tag (see Supplemental Figure 1B online). The protein was purified by metal affinity chromatography using Ni-NTA. Activity of the enzyme was assayed in 50 mM Tris-HCl buffer, pH 7.5, containing 4 mM MnCl2, 0,2 mg/mL BSA, 1 mM GTP, 1 mM IBMX, 1% DMSO, and the enzyme. The assay was performed at 37°C for 10 min. Determination of cGMP was performed according to the immunoassay method (cGMP enzyme immuno assay kit direct; Sigma-Aldrich).
Determination of NH4+ Concentration
NH4+ concentration was determined using the Nessler reagent by adding 25 μL of reagent A and 25 μL of reagent B to 500 μL medium and immediate measurement of OD at λ = 410 nm (ϵ = 1.67 mM−1 cm−1) according to Solórzano (1969).
Primers and PCR
All primers were designed using Primer Select (DNA Star version 4. 05). For all the primer pairs used in the real-time PCR experiments, at least one of the two primers was designed so that its sequence would be overlapping two exons to avoid DNA contamination effects. Moreover, for each primer pair, the PCR product was run on a gel and sequenced to be sure of its specificity. Optimization of the primers was performed with an annealing temperature gradient and DMSO concentration gradient. In the real-time PCR experiments, the housekeeping gene corresponds to a ubiquitin ligase (EST references: BU648530, BU648531, BE237749, and BE237718) that shows constitutive expression under all the experimental conditions (González-Ballester et al., 2004). Primer sequences are given in Table 1.
Table 1.
List of primers used
| Primer | Sequence (5′ to 3′) | Use |
| Actinup | ACTGAGCACAATGTTAC | Arabidopsis ACTIN gene |
| Actinlw | GGTGATGGTGTGTCT | |
| AtNia1up | AGGCTACGCTTATTCTGGAGGA | Arabidopsis NIA1 |
| AtNia1lw | GAACCGCGACGTCTTTAGCACTGAGCA | |
| AtNia2up | CGCCGACGAAGAAGGTTGG | Arabidopsis NIA2 |
| AtNia2lw | GGGTTGTGAAAGCGTTGATGGGAAGAAT | |
| Ubiupper | GTACAGCGGCGGCTAGAGGCAC | Chlamydomonas (Cr) gene for ubiquitin |
| Ubilower | AGCGTCAGCGGCGGTTGCAGGTATCT | |
| 20.73RT1fw | GAGCGCTTCGAGGCAGAGACC | Cr CYG56 transcript |
| 20.73RT1rev | CAGCAGTGCGTCAAACTCGTTGTACA | |
| NRupper | CCGAGCGCTTCCGGCTGTGGTACA | Cr NIA1 transcript |
| NRlower | CTGGATCTGGCGGTCCTTGCTGTA | |
| Nrt2.1upper | CGCCGTGGCAACTGACCCTGAG | Cr NRT2.1 transcript |
| Nrt2.1lower | CGCCACCTCCTCCGCACTCCAC | |
| AMT1.1upper | GCACGGGAGGGCAAGAGGTTC | Cr AMT1.1 transcript |
| AMT1.1lower | ATGTGCCGCAGTCAAGAAGGATTT | |
| AMT1.2upper | GGCTCGCCACCTGCAAGAGACAAC | Cr AMT1.2 transcript |
| AMT1.2lower | GATGGCACCGGCCTCGAGCATAGC | |
| Primer 1 | ATGCAATTTCTTCTAGATGAACCCGT | see Figure 3 |
| Primer 2 | CCAGAGCTGCCACCTTGACA | |
| Primer 3 | CCACCACCCCGAAGCCGATAA | |
| Primer 4 | TCCTCCACAACAACCCACTCACAACC | |
| Primer 5 | CGCCCTCATAGCCCGCCAAAT | |
| Primer 6 | TCCCGCTGCTTCTTGCTGCT | |
| Primer 7 | CAGCCCCCAGGATCTTGTA |
Chemicals
Adequate amounts of products were weighed on the day of each experiment and resuspended just before application. DEA NONOate was resuspended in minimum medium for Chlamydomonas and liquid MS medium for Arabidopsis, and the pH of the solutions was adjusted to 7.2. After resuspension, DEA NONOate was applied immediately. In the experiment shown in Figure 11, 200 μL of DEA NONOate 100 μM was sprayed directly on the seedlings; the small size of the wells (2.5 cm diameter) allowed the liquid to stay in contact with the plants. Independent treatments were done in each well.
Isolation of RNA and cDNA Synthesis
Chlamydomonas cells were grown in minimum medium with 8 mM NH4Cl, collected at mid-exponential phase of growth (4000g, 1 min) and transferred to medium containing the specified nitrogen source. RNA was extracted after lysis of the cells with 2% SDS in 100 mM Tris-HCl, pH 8.0, 400 mM NaCl, and 50 mM EDTA. Total RNA was extracted with phenol/chloroform as previously reported (Schloss et al., 1984).
Arabidopsis tissue was harvested after 10 d of growth in the previously described conditions. RNA was isolated from whole seedlings using the Qiagen RNeasy extraction kit, following the recommendations of the manufacturer. Chlamydomonas and Arabidopsis RNA was reversed transcribed using oligo(dT) primers and Superscript II reverse transcriptase (Invitrogen).
RT and Real-Time PCR
RT-PCR reactions were made in a 25-μL final volume with the following components: 0.2 pmol of each primer, 0.2 mM deoxynucleotide triphosphate, 0.5 units Taq DNA polymerase (BandM Labs), 2 mM MgCl2, 1 μL cDNA (synthesized from 3 μg of total RNA), and 2.5 μL of the specific buffer. The standard thermal profile was 95°C, 5 min; 40× (95°C, 30 s; 60°C, 30 s; 72°C, 5 min). Real-time PCR was performed using the LightCycler Instrument (iCycler iQ real-time PCR detection system; Bio-Rad) with SYBR Green I (Molecular Probes) as a fluorescent dye. Each individual reaction was made in a 25-μL final volume with the same components as for RT-PCR complemented with 1.25 μL of SYBR Green diluted 10−4 in DMSO. The LightCycler-run protocol was 95°C, 5 min; 40× (95°C, 30 s; 60°C, 30 s; 72°C, 15 s), and fluorescence measurements were done at 84°C for 10 s. The specificity of the PCR products was checked by a melting curve program and analyzed on a 2% agarose gel electrophoresis. The amplification rate of each transcript (Ct) was calculated by the PCR baseline subtracted method performed with LightCycler software (iCycler iQ, Optical System Software, version 3) at a constant fluorescence level. Cts were determined over three repeats within the LightCycler and with three different runs.
DNA Sequencing and Sequence Analysis
DNA sequencing was performed at the Servicio Central de Apoyo a la Investigación (University of Córdoba, Spain) with fluorescence-labeled terminators for an automatic sequencer (model ABI 377; Applied Biosystems, Perkin-Elmer) according to the manufacturer's instructions. Sequences were analyzed using the DNAstar software v. 4.05 (Lasergene Navigator), the Bioedit Sequence Alignment Editor v. 5.0.9 (Department of Microbiology, North Carolina State University), the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/), and the Chlamydomonas Joint Genome Initiative server (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). Domain predictions were analyzed with Expasy (http://www.expasy.ch/cgi-bin/prosite/ and http://myhits.isb-sib.ch/cgi-bin/motif_scan) and Smart (http://smart.embl-heidelberg.de/) servers. Clustal comparisons were performed with ClustalW from the BioEdit program. Similarity scores were obtained with MegAlign from the DNASTAR package.
NO Quantification
Chlamydomonas cells were grown in minimum medium with 8 mM NH4Cl, collected at mid-exponential phase of growth, and transferred to medium containing different nitrogen sources. DAF-FM DA (1 μM; Invitrogen Molecular Probes) (Kojima et al., 1998) was applied 30 min prior to the measurement. Cells were then washed and resuspended in 100 mM HEPES buffer, pH 7.5. Cells (200 μL) were transferred to a black optiplate 96F (Perkin-Elmer Life Sciences), and fluorescence was measured in a Multidetection microplate reader (Synergy 4; Biotek). Excitation and emission wavelengths were 495 and 515 nm, respectively. Three to eight technical replicates per condition were included on each plate.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers. Sequences from Chlamydomonas were as follows: CYG56 cDNA, EU841916; CYG56, ACJ05390.1; NIA1, AF203033; NRT2.1, Z25438; AMT1.1, AF479643; and AMT1.2, AF530051. Sequences from Arabidopsis were as follows: NIA1, P11832; and NIA2, P11035.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genomic Structure of CYG56 and Domains of the CYG56 Protein.
Supplemental Figure 2. Sequence Comparison Annealing of HNOB and GC Domains.
Supplemental Figure 3. Sequence Comparison Annealing of HNOBA Domain.
Supplemental Figure 4. Degree of Identity and Divergence among Compared NO-Dependent Type GC Sequences.
Supplemental Figure 5. GC Activity Assayed with Mg2+ and Mn2+ of Truncated (pCYG56) and Full-Length CYG56 Purified Preparations.
Supplemental Figure 6. Effect of the Inhibitors ODQ and LY83,583 on GC Activity of Truncated and Full-Length CYG56 Purified Preparations.
Supplemental Figure 7. Effect of 8-Br-cGMP on Expression of NIA1, NRT2.1, AMT1.1, and AMT1.2 in the Wild-Type 704.
Supplemental Figure 8. DAF-FM DA Fluorescence Is Proportional to NO Intracellular Levels.
Supplemental Table 1. Raw Data Used to Produce Figure 10.
Supplementary Material
Acknowledgments
We thank Maribel Macías for skillful technical assistance and A. Llamas for his help in protein expression. E.S.-L. thanks the Ministerio de Educación y Ciencia, Spain, for a PhD fellowship. This work was funded by the Ministerio de Educación y Ciencia (Grants BFU2005-07521 and BFU2008-01798), by the European Commission (INTAS, Grant 10000008-8004), and by the Junta de Andalucía, Spain (PAI, CVI-0128).
References
- Ali R., Ma W., Lemtiri-Chlieh F., Tsaltas D., Leng Q., von Bodman S., Berkowitz G.A. (2007). Death don't have no mercy and neither does calcium: Arabidopsis cyclic nucleotide gated channel2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D.I. (1949). Copper enzymes in isolated chloroplasts: Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke P.C., Badger M.R., Jones R.L. (2004). Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 16: 332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke P.C., Libourel I.G., Reinohl V., Jones R.L. (2005). Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta 223: 805–812 [DOI] [PubMed] [Google Scholar]
- Boss G.R. (1989). cGMP-induced differentiation of the promyelocytic cell line HL-60. Proc. Natl. Acad. Sci. USA 86: 7174–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büsch A., Strube K., Friedrich B., Cramm R. (2005). Transcriptional regulation of nitric oxide reduction in Ralstonia eutropha H16. Biochem. Soc. Trans. 33: 193–194 [DOI] [PubMed] [Google Scholar]
- Camargo A., Llamas A., Schnell R.A., Higuera J.J., González-Ballester D., Lefebvre P.A., Fernández E., Galván A. (2007). Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell 19: 3491–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaigns L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., Taconnat L., Renou J.P., Daniel-Vedele F., Fernandez E., Meyer C., Krapp A. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57: 426–435 [DOI] [PubMed] [Google Scholar]
- Corpas F.J., Barroso J.B., Carreras A., Quirós M., León A.M., Romero-Puertas M.C., Esteban F.J., Valderrama R., Palma J.M., Sandalio L.M., Gómez M., del Río L.A. (2004). Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 136: 2722–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N.M. (1995). Nitrate: Nutrient and signal for plant growth. Plant Cell 7: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N.M., Forde B.G. (2002). Molecular and developmental biology of inorganic nitrogen nutrition. In The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., eds (Rockville, MD: American Society of Plant Biologists; ), doi/, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois C., Besson A., Dahan J., Bourque S., Dobrowolska G., Pugin A., Wendehenne D. (2008). Nitric oxide signalling in plants: Interplays with Ca2+ and protein kinases. J. Exp. Bot. 59: 155–63 [DOI] [PubMed] [Google Scholar]
- Cueto M., Hernandez-Perera O., Martin R., Bentura M.L., Rodrigo J., Lamas S., Golvano M.P. (1996). Presence of nitric oxide synthase activity in roots and nodules of Lupinus albus. FEBS Lett. 398: 159–164 [DOI] [PubMed] [Google Scholar]
- Daniel-Vedele F., Filleur S., Caboche M. (1998). Nitrate transport: A key step in nitrate assimilation. Curr. Opin. Plant Biol. 1: 235–239 [DOI] [PubMed] [Google Scholar]
- Dean J.V., Harper J.E. (1986). Nitric oxide and nitrous oxide production by soybean and winged bean during the in vivo nitrate reductase assay. Plant Physiol. 82: 718–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J.V., Harper J.E. (1988). The conversion of nitrite to nitrogen oxide(s) by the constitutive NAD(P)H-nitrate reductase enzyme from soybean. Plant Physiol. 88: 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M., Xia Y., Dixon R.A., Lamb C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- del Rio L.A., Corpas F.J., Barroso J.B. (2004). Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry 65: 783–792 [DOI] [PubMed] [Google Scholar]
- Douglas P., Pigaglio E., Ferrer A., Halfords N.G., MacKintosh C. (1997). Three spinach leaf nitrate reductase-3-hydroxy-3-methylglutaryl-CoA reductase kinases that are required by reversible phosphorylation and/or Ca2+ ions. Biochem. J. 325: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J., Wendehenne D., Klessig D.F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Gordon-Weeks R., Shen Q., Miller A.J. (2006). Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J. Exp. Bot. 57: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Fernandez E., Cardenas J. (1982). Regulation of the nitrate-reducing system enzymes in wild type and mutant strains of Chlamydomonas reinhardtii. Mol. Gen. Genet. 186: 164–169 [DOI] [PubMed] [Google Scholar]
- Fernandez E., Galvan A. (2007). Inorganic nitrogen assimilation in Chlamydomonas. J. Exp. Bot. 58: 2279–2287 [DOI] [PubMed] [Google Scholar]
- Fernandez E., Galvan A., Quesada A. (1998). Nitrogen assimilation and its regulation. Molecular Biology of Chlamydomonas: Chloroplast and Mitochondria, Rochaix J.D., Goldschmidt-Clermont M., Merchant S., eds (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 637–659 [Google Scholar]
- Fernandez E., Matagne R.F. (1984). Genetic analysis of nitrate reductase-deficient mutants in Chlamydomonas reinhardtii. Curr. Genet. 8: 635–640 [DOI] [PubMed] [Google Scholar]
- Fernández E., Matagne R.F. (1986). In vivo complementation analysis of nitrate reductase-deficient mutants in Chlamydomonas reinhardtii. Curr. Genet. 10: 397–403 [DOI] [PubMed] [Google Scholar]
- Fernhoff N.B., Derbyshire E.R., Marletta M.A. (2009). A nitric oxide/cysteine interaction mediates the activation of soluble guanylate cyclase. Proc. Natl. Acad. Sci. USA 106: 21602–21607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B.G. (2000). Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Galangau F., Daniel-Vedele F., Moureaux T., Dorbe M.F., Leydecker M.T., Caboche M. (1998). Expression of leaf nitrate reductase genes from tomato and tobacco in relation to light-dark regimes and nitrate supply. Plant Physiol. 88: 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván A., González-Ballester D., Fernandez E. (2007). Insertional mutagenesis as a tool to study genes/functions in Chlamydomonas. Adv. Exp. Med. Biol. 616: 77–89 [DOI] [PubMed] [Google Scholar]
- Galván A., Mariscal V., González-Ballester D., Fernández E. (2006). The green alga Chlamydomonas as a tool to study the nitrate assimilation pathway in plants. In Model Plants and Crop Improvement, Varshney R.K., Koebner R.M.D., eds (Boca Raton, FL: CRC Press; ), pp. 125–158 [Google Scholar]
- Glass A.D.M. (2003). Nitrogen use efficiency of crop plants: Physiological constraints upon nitrogen absortion. Crit. Rev. Plant Sci. 22: 453–470 [Google Scholar]
- González-Ballester D., Camargo A., Fernandez E. (2004). Ammonium transporter genes in Chlamydomonas: The nitrate-specific regulatory gene Nit2 is involved in AMT1;1 expression. Plant Mol. Biol. 56: 863–878 [DOI] [PubMed] [Google Scholar]
- González-Ballester D., de Montaigu A., Galván A., Fernández E. (2005b). Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal. Biochem. 340: 330–335 [DOI] [PubMed] [Google Scholar]
- González-Ballester D., de Montaigu A., Higuera J.J., Galvan A., Fernandez E. (2005a). Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol. 137: 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea C.M.C.P., Souza J.F., Magalhaes A.C.N., Martins I.S. (1997). NO-releasing substances that induce growth elongation in maize root segments. Plant Growth Regul. 21: 183–187 [Google Scholar]
- Guo F.Q., Okamoto M., Crawford N.M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- Harris E. (1989). The Chlamydomonas Sourcebook. (New York: Academic Press; ). [Google Scholar]
- He Y., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Ignarro L.J., Fukuto J.M., Griscavage J.M., Rogers N.E., Byrns R.E. (1993). Oxidation of nitric oxide in aqueous solution to nitrite but not nitrate: comparison with enzymatically formed nitric oxide from L-arginine. Proc. Natl. Acad. Sci. USA 90: 8103–8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Anantharaman V., Aravind L. (2003). Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C.W., Du S.T., Zhang Y.S., Lin X.Y., Tang C.X. (2009). Differential regulatory role of nitric oxide in mediating nitrate reductase activity in roots of tomato (Solanum lycocarpum). Ann. Bot. (Lond.) 104: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser W.M., Brendle-Behnisch E. (1991). Rapid modulation of spinach leaf nitrate reductase activity by photosynthesis: I. Modulation in vivo by CO2 availability. Plant Physiol. 96: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser W.M., Huber S.C. (2001). Post-translational regulation of nitrate reductase: Mechanism, physiological relevance and environmental triggers. J. Exp. Bot. 52: 1981–1989 [DOI] [PubMed] [Google Scholar]
- Koglin M., Vehse K., Budaeus L., Scholz H., Behrends S. (2001). Nitric oxide activates the β2 subunit of soluble guanylyl cyclase in the absence of a second subunit. J. Biol. Chem. 276: 30737–30743 [DOI] [PubMed] [Google Scholar]
- Kojima H., Nakatsubo N., Kikuchi K., Kawahara S., Kirino Y., Nagoshi H., Hirata Y., Nagano T. (1998). Detection and imaging of nitric oxide with novel fluorescent indicators: Diaminofluoresceins. Anal. Chem. 70: 2446–2453 [DOI] [PubMed] [Google Scholar]
- Kozak M. (2002). Pushing the limits of the scanning mechanism for initiation of translation. Gene 299: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwezi L., Meier S., Mungur L., Ruzvidzo O., Irving H., Gehring C. (2007). The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS One 23: e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L., Garcia-Mata C., Graziano M., Pagnussat G. (2003). Nitric oxide: The versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54: 109–136 [DOI] [PubMed] [Google Scholar]
- Lanteri M.L., Pagnussat G.C., Lamattina L. (2006). Calcium and calcium-dependent protein kinases are involved in nitric oxide- and auxin-induced adventitious root formation in cucumber. J. Exp. Bot. 57: 1341–1351 [DOI] [PubMed] [Google Scholar]
- Lea U.S., Ten Hoopen F., Provan F., Kaiser W.M., Meyer C., Lillo C. (2004). Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in high nitrite excretion and NO emission from leaf and root tissue. Planta 219: 59–65 [DOI] [PubMed] [Google Scholar]
- Lillo C., Lea U.S., Leydecker M.T., Meyer C. (2003). Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in constitutive activation of the enzyme in vivo and nitrite accumulation. Plant J. 35: 566–573 [DOI] [PubMed] [Google Scholar]
- Llamas A., Igeno M.I., Galvan A., Fernandez E. (2002). Nitrate signaling on the nitrate reductase gene promoter depends directly on the activity of the nitrate transport systems in Chlamydomonas. Plant J. 30: 261–271 [DOI] [PubMed] [Google Scholar]
- Loppes R., Ohresser M., Radoux M., Matagne R.F. (1999). Transcriptional regulation of the Nit1 gene encoding nitrate reductase in Chlamydomonas reinhardtii: Effect of various enviromental factors on the expression of a reporter gene under the control of the Nit1 promoter. Plant Mol. Biol. 41: 701–711 [DOI] [PubMed] [Google Scholar]
- Loque D., Tillard P., Gojon A., Lepetit M. (2003). Gene expression of the NO3- transporter NRT1.1 and the nitrate reductase NIA1 is repressed in Arabidopsis roots by NO2-, the product of NO3- reduction. Plant Physiol. 132: 958–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludidi N., Gehring C. (2003). Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J. Biol. Chem. 278: 6490–6494 [DOI] [PubMed] [Google Scholar]
- Ma X., Sayed N., Baskaran P., Beuve A., van den Akker F. (2008). PAS-mediated dimerization of soluble guanylyl cyclase revealed by signal transduction histidine kinase domain crystal structure. J. Biol. Chem. 283: 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis F.J. (2006). cGMP modulates gene transcription and cation transport in Arabidopsis roots. Plant J. 45: 700–711 [DOI] [PubMed] [Google Scholar]
- Mata C.G., Lamattina L. (2001). Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol. 126: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M., Lee G.I., Wang Y., Crane B.R., Klessig D.F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283: 32957–32967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nioche P., Berka V., Vipond J., Minton N., Tsai A.L., Raman C.S. (2004). Femtomolar sensitivity of a NO sensor from Clostridium botulinum. Science 306: 1550–1553 [DOI] [PubMed] [Google Scholar]
- Oh S., McCaslin P.P. (1995). The iron component of sodium nitroprusside blocks NMDA-induced glutamate accumulation and intracellular Ca2+ elevation. Neurochem. Res. 20: 779–784 [DOI] [PubMed] [Google Scholar]
- Ohresser M., Matagne R., Loppes R. (1997). Expression of the arylsulphatase reporter gene under the control of the nit1 promoter in Chlamydomonas reinhardtii. Curr. Genet. 31: 264–271 [DOI] [PubMed] [Google Scholar]
- Pellicena P., Karow D.S., Boon E.M., Marletta M.A., Kuriyan J. (2004). Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc. Natl. Acad. Sci. USA 101: 12854–12859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Alegre M., Dubus A., Fernandez E. (2005). REM1, a new type of long terminal repeat retrotransposon in Chlamydomonas reinhardtii. Mol. Cell. Biol. 25: 10628–10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchet E., Gupta J.K., Sonoda M., Kaiser W.M. (2005). Nitric oxide emission from tobacco leaves and cell suspensions: rate-limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 41: 732–743 [DOI] [PubMed] [Google Scholar]
- Planchet E., Kaiser W.M. (2006). Nitric oxide (NO) detection by DAF fluorescence and chemiluminescence: a comparison using abiotic and biotic NO sources. J. Exp. Bot. 57: 3043–3055 [DOI] [PubMed] [Google Scholar]
- Pozuelo M., MacKintosh C., Galván A., Fernández E. (2001). Cytosolic glutamine synthetase and not nitrate reductase from the green alga Chlamydomonas reinhardtii is phosphorylated and binds 14-3-3 proteins. Planta 212: 264–269 [DOI] [PubMed] [Google Scholar]
- Prieto R., Dubus A., Galván A., Fernández E. (1996). Isolation and characterization of two regulatory mutants for nitrate assimilation in Chlamydomonas reinhardtii. Mol. Gen. Genet. 251: 461–471 [DOI] [PubMed] [Google Scholar]
- Radomski M.W., Palmer R.M., Moncada S. (1990). Characterization of the L-arginine:nitric oxide pathway in human platelets. Br. J. Pharmacol. 101: 325–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach J., Fernandez E., Galvan A. (2000). The Chlamydomonas reinhardtii Nar1 gene encodes a chloroplast membrane protein involved in nitrite transport. Plant Cell 12: 1441–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach J., Llamas A., Fernandez E., Galvan A. (2002). The activity of the high-affinity nitrate transport system I (NRT2;1, NAR2) is responsible for the efficient signaling of nitrate assimilation genes in Chlamydomonas reinhardtii. Planta 215: 606–611 [DOI] [PubMed] [Google Scholar]
- Rockel P., Strube F., Rockel A., Wildt J., Kaiser W.M. (2002). Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53: 103–110 [PubMed] [Google Scholar]
- Sakihama Y., Nakamura S., Yamasaki H. (2002). Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: An alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol. 43: 290–297 [DOI] [PubMed] [Google Scholar]
- Salmi M.L., Morris K.E., Roux S.J., Porterfield D.M. (2007). Nitric oxide and cGMP signaling in calcium-dependent development of cell polarity in Ceratopteris richardii. Plant Physiol. 144: 94–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss J.A., Silflow C.D., Rosenbaum J.L. (1984). mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol. Cell. Biol. 4: 424–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell R.A., Lefebvre P.A. (1993). Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C.D. (1998). Organization of the nuclear genome. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, Rochaix J.-D., Goldsmidt-Clermont M., Merchant S., eds (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 25–40 [Google Scholar]
- Slocum R.D. (2005). Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 43: 729–745 [DOI] [PubMed] [Google Scholar]
- Snell F.D., Snell C.T. (1949). Colorimetric Methods of Analysis, Vol. 2. (New York: Van Nostrand; ). [Google Scholar]
- Sokolovski S., Hills A., Gay R., Garcia-Mata C., Lamattina L., Blatt M.R. (2005). Protein phosphorylation is a prerequisite for intracellular Ca2+ release and ion channel control by nitric oxide and abscisic acid in guard cells. Plant J. 43: 520–529 [DOI] [PubMed] [Google Scholar]
- Solórzano L. (1969). Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol. Oceanogr. 14: 799–801 [Google Scholar]
- Stitt M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Tischner R., Planchet E., Kaiser W.M. (2004). Mitochondrial electron transport as a source for nitric oxide in the unicellular green alga Chlorella sorokiniana. FEBS Lett. 576: 151–155 [DOI] [PubMed] [Google Scholar]
- von Gromoff E.D., Schroda M., Oster U., Beck C.F. (2006). Identification of a plastid response element that acts as an enhancer within the Chlamydomonas HSP70A promoter. Nucleic Acids Res. 34: 4767–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H., Sakihama Y. (2000). Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: In vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 468: 89–92 [DOI] [PubMed] [Google Scholar]
- Yang Q.Z., Hatton G.I. (1999). Nitric oxide via cGMP-dependent mechanisms increases dye coupling and excitability of rat supraoptic nucleus neurons. J. Neurosci. 19: 4270–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Ali M.L., Taylor J., Liu J., Sun G., Liu W., Masilimany P., Gulati-Sakhuja A., Pauls K.P. (2008). A guanylyl cyclase-like gene is associated with Gibberella ear rot resistance in maize (Zea mays L.). Theor. Appl. Genet. 116: 465–479 [DOI] [PubMed] [Google Scholar]
- Zemojtel T., Frohlich A., Palmieri M.C., Kolanczyk M., Mikula I., Wyrwicz L.S., Wanker E.E., Mundlos S., Vingron M., Martasek P., Durner J. (2006). Plant nitric oxide synthase: A never-ending story? Trends Plant Sci. 11: 524–525 [DOI] [PubMed] [Google Scholar]
- Zhang D., Lefebvre P.A. (1997). Far1, a negative regulatory locus required for the repression of the nitrate reductase gene in Chlamydomonas reinhardtii. Genetics 146: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Brandish P.E., DiValentin M., Schelvis J.P.M., Babcock G.T., Marletta M.A. (2000). Inhibition of soluble guanylate cyclase by ODQ. Biochemistry 39: 10848–10854 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.