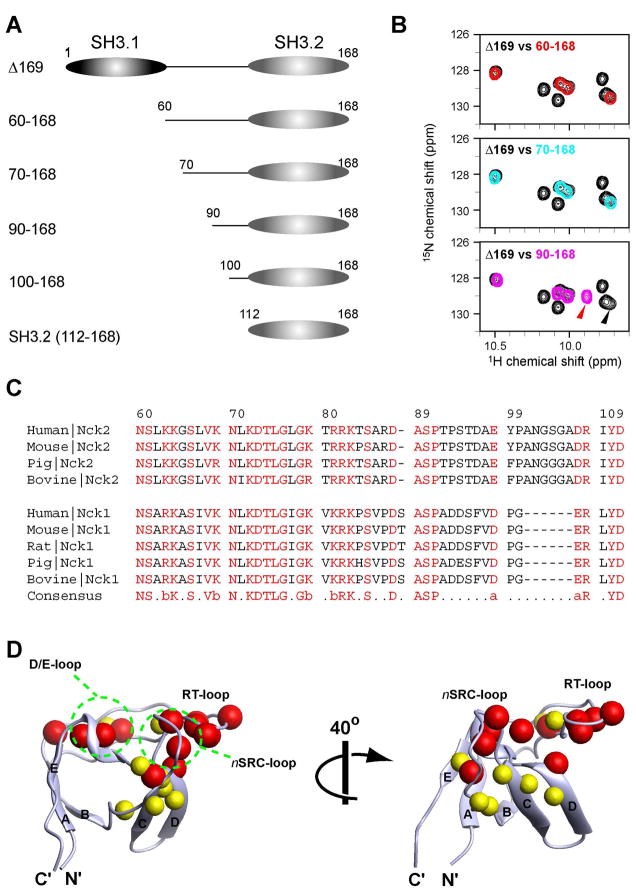
Figure 3.
N, C-terminal deletion constructs of Nck2 (A) Schematic representation of the N, C-terminal deletion constructs of Nck2 (B) Comparison of 1H15N HSQC spectrum between 100 μM [ul-15N] Nck2 Δ169 and 100 μM [ul-15N] Nck2_60–168 (top, red contour), [ul-15N] Nck2_70–168 (middle, cyan contour), or [ul-15N] Nck2_90–168 (middle, pink contour). The region containing the tryptophane indole signals is shown. The peak positions of W148 indole, which shifted significantly between Nck2 Δ169 and Nck2_90–168, are indicated with arrowheads that have same color with its spectrum. (C) Sequence alignment of human, mouse, rat, pig and bovine Nck proteins at the SH3.1/SH3.2 linker regions. (D) Mapping of the residues that are shifted by the deletion of the residues 70–89 (red) or 90–110 (yellow). Residues that are shifted >0.05 ppm in Nck2_70–168 with respect to 90–168 are indicated as red spheres. Yellow spheres are for the residues shifted >0.05 ppm in Nck2_90–168 with respect to isolated SH3.2. Residues shifted >0.05 ppm in both cases are indicated with red spheres. Chemical shift changes are normalized by the equation :((δN/5)2+(δH)2)1/2, where δN and δH are chemical shit changes in nitrogen and proton dimensions. Although Asn112 was also shifted >0.05 ppm in Nck2_90–168 with respect to isolated SH3.2, the residue is not shown in Figure 4D since it is not defined in the determined structure. The solution structure of the isolated SH3.2 domain, which contains residues 114–170 (2FRW), was used for mapping (16).