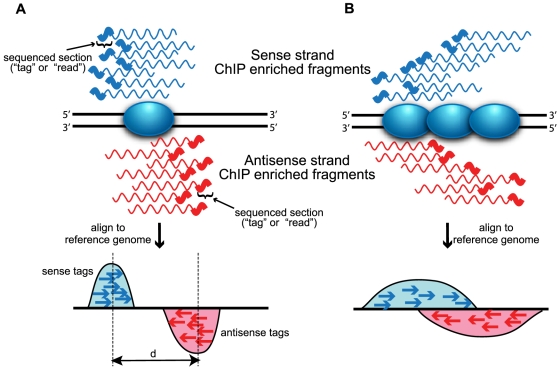
Figure 1. Strand-dependent bimodality in tag density.
The 5′ to 3′ sequencing requirement and short read length produce stranded bias in tag distribution. The shaded blue oval represents the protein of interest bound to DNA (solid black lines). Wavy lines represent either sense (blue) or antisense (red) DNA fragments from ChIP enrichment. The thicker portion of the line indicates regions sequenced by short read sequencing technologies. Sequenced tags are aligned to a reference genome and projected onto a chromosomal coordinate (red and blue arrows). (A) Sequence-specific binding events (e.g. transcription factors) are characterized by “punctuate enrichment” [11] and defined strand-dependent bimodality, where the separation between peaks (d) corresponds to the average sequenced fragment length. Panel A was inspired by Jothi et al. [32]. (B) Distributed binding events (e.g. histones or RNA polymerase) produce a broader pattern of tag enrichment that results in a less defined bimodal pattern.