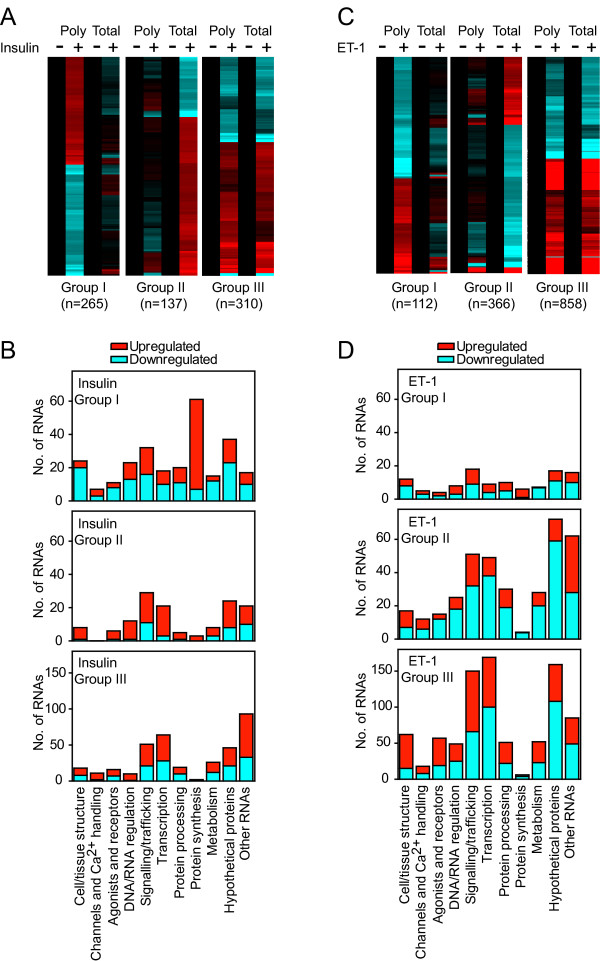
Figure 4.
Translational regulation of cardiomyocyte transcripts by insulin or ET-1.Cardiomyocytes were exposed to 100 nM ET-1 or 50 mU/ml insulin (1 h) or were left unstimulated. Polysomes were prepared by sucrose density centrifugation. Total and polysomal RNA were extracted and analysed using microarrays. RNAs with significant changes in expression (FDR < 0.05, >1.25-fold change) induced by insulin (A and B) or ET-1 (C and D) in either total or polysomal RNA pools were identified and clustered according to regulation in polysomal RNA only (Group I; FDR < 0.05 in polysomal RNA only and expression ratio >1.2 for polysomal RNA:total RNA), regulation in total RNA only (Group II; FDR < 0.05 in total RNA only and expression ratio >1.2 for total RNA:polysomal RNA) or similar regulation in both (Group III; FDR < 0.05 in both polysomal and total RNA, or expression ratio <1.2). A, Heatmap for insulin-regulated probesets showing the mean values of all probesets in each group with normalisation per gene to control values [Log2 scale; -0.7 (cyan) through 0 (black) to 0.7 (red)]. C, Heatmap for ET-1-regulated probesets showing the mean values of all probesets in each group with normalisation per gene to control values [Log2 scale; -1.5 (cyan) through 0 (black) to 1.5 (red)]. B and D, Functional classification of the RNAs illustrated in panels A and C, respectively, showing the numbers of RNAs in each group (numbers of upregulated and downregulated RNAs are represented in red and cyan, respectively).