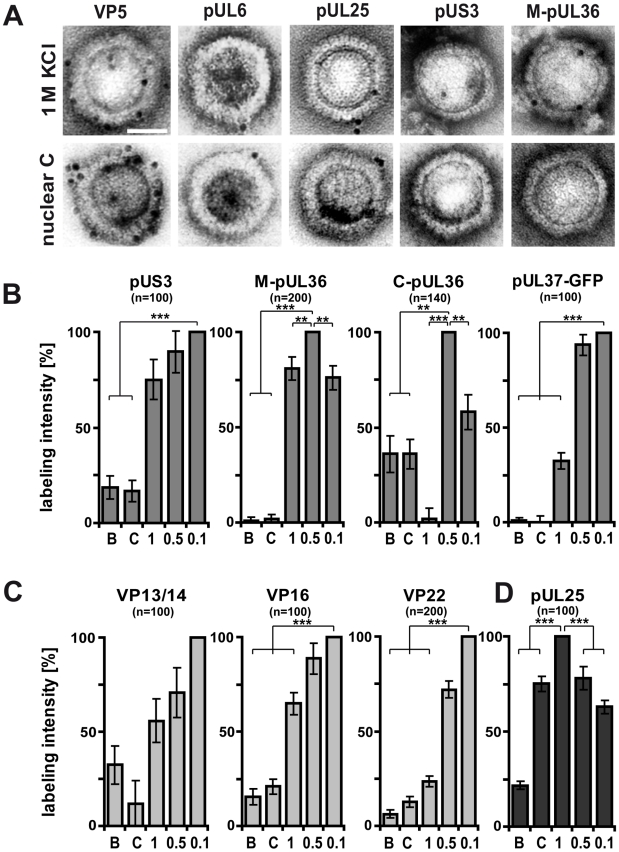
Figure 8. Generation of different HSV1 capsid surfaces during isolation.
HSV1(F) capsids were isolated from infected nuclei (B or C) or prepared from extracellular virions by detergent lysis in the presence of different KCL concentrations (1, 0.5 or 0.1 M KCl), and labeled with antibodies against the capsid proteins VP5 (A: pAb NC-1), pUL25 (A, D: pAb ID1) or pUL6 (A: mAb 1C), against the inner tegument proteins pUS3, (A: pAb anti-US3), pUL36 (A, B: pAb #147, anti-middle-pUL36; B: pAb anti-Cterminal-pUL36), pUL37-GFP (B: mAb anti-GFP JL-8; here capsids from strain HSV1-pUL37GFP), or the outer tegument proteins VP13/14 (C: pAb R220), VP16 (D: pAb SW7), or VP22 (D: pAb AGV30). A: Immunoelectron microscopy images of capsids after immunolabeling followed by protein-A gold and negative staining. Scale bar: 50 nm. B to D: The labeling intensity for inner tegument proteins (B), outer tegument proteins (C) or the capsid associated protein pUL25 (D) was quantified by counting the number of gold particles per capsid. After subtraction of the background without the primary antibodies, the number on the capsid type with the highest labeling was set to 100%, and recalculated accordingly for the other capsid types. These data sets were also directly integrated into Fig. 3 (red columns). Error bars: SEM. n: number of capsids (same for each capsid type with a given antibody) summarized from three experiments. Two asterisks denote p<0.001 and three asterisks indicate P<0.0001 as determined in two-sided Student's t-tests.