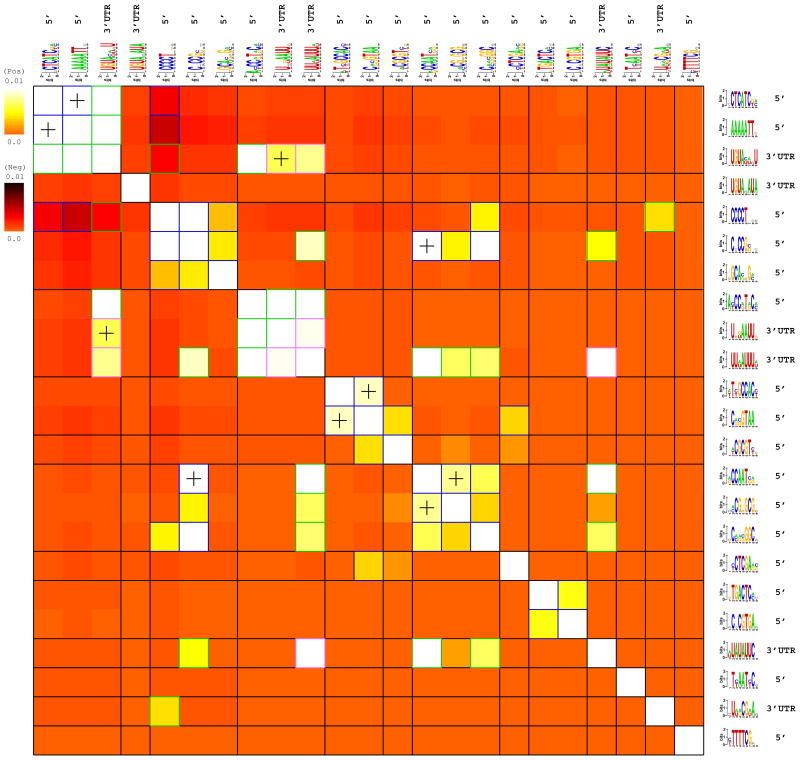
Figure 4. Interactions among all predicted yeast motifs.
Each row/column corresponds to a predicted motif. The color-map indicates the level by which the presence of one motif implies the presence (light color-map) or the absence (dark color-map) of another motif within the same promoter, as quantified by their interaction information (see Supplementary Methods). Very light colors indicate strong positive co-occurrences between pairs of motifs that have further been used to construct putative functional modules, indicated on the figure (and in Figure 3). Very dark colors indicate that the two motifs tend to avoid being present within the same promoter. Statistically significant information values (p<1e-4) that involve homotypic motif pairs are highlighted using blue (DNA-DNA) and pink (RNA-RNA) frames, while those that involve heterotypic pairs (a DNA motif and a RNA motif) are highlighted using green frames. Significant spatial co-localization between pairs of motifs are denoted with ‘+’. For more details, see the Supplementary Methods section about FIRE interaction heat-maps.