Summary
AMP-activated protein kinase (AMPK) is a master metabolic regulator for controlling cellular energy homeostasis. Its homolog in yeast, SNF1, is activated in response to glucose depletion and other stresses. The catalytic (α) subunit of AMPK/SNF1, Snf1 in yeast, contains a protein Ser/Thr kinase domain (KD), an auto-inhibitory domain (AID), and a region that mediates interactions with the two regulatory (β and γ) subunits. Previous studies suggested that Snf1 contains an additional segment, a regulatory sequence (RS, corresponding to residues 392-518), which may also have an important role in regulating the activity of the enzyme. The crystal structure of the heterotrimer core of S. cerevisiae SNF1 showed interactions between a part of the RS (residues 460-498) and the γ subunit Snf4. Here we report biochemical and functional studies on the regulation of SNF1 by the RS. GST pulldown experiments demonstrate strong and direct interactions between residues 450-500 of the RS and the heterotrimer core, and single-site mutations in the RS-Snf4 interface can greatly reduce these interactions in vitro. On the other hand, functional studies appear to show only small effects of the RS-Snf4 interactions on the activity of SNF1 in vivo. This suggests that residues 450–500 may be constitutively associated with Snf4, and the remaining segments of the RS, as well as the AID, may be involved in regulating SNF1 activity.
Keywords: fatty acid metabolism, metabolic syndrome, protein kinase, auto-inhibitory domain (AID)
Introduction
AMP-activated protein kinase (AMPK) is a central player in cellular responses to low energy levels, and is an attractive drug discovery target against type 2 diabetes, obesity and other human diseases [1–4]. The S. cerevisiae homolog of AMPK, known as Sucrose Non-Fermenting 1 (SNF1) [1], has similar functions as the higher eukaryotic AMPKs [5]. AMPK/SNF1 proteins are conserved, heterotrimeric (αβγ) enzymes in most eukaryotes [1–4]. The α subunit consists of an N-terminal protein Ser/Thr kinase domain (KD) [6; 7], followed by an auto-inhibitory domain (AID) [8–10] and a region for interactions with the other two subunits (Fig. 1A). The α subunit of SNF1, known as Snf1, also contains a regulatory sequence (RS) just after the AID in the primary sequence (Fig. 1A, and see below) [11]. The β subunit serves as the scaffold of the heterotrimer, with a C-terminal αγ interacting region [12; 13] and, immediately N-terminal to it, a glycogen binding domain (GBD) [14–16], now also known as the carbohydrate binding module (CBM) [17; 18]. The γ subunit contains four cystathionine β-synthase (CBS) motifs, each tandem pair of which is called a Bateman domain [19; 20]. Each Bateman domain is capable of binding two molecules of AMP or ATP [21–23], allowing AMPK to sense cellular energy levels.
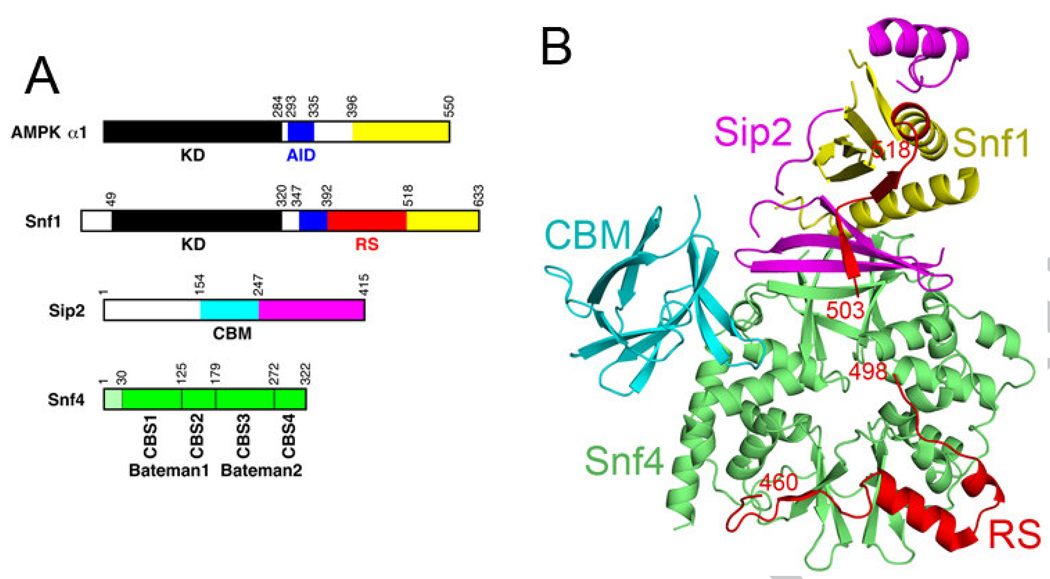
Figure 1. Structure of the S. cerevisiae SNF1 heterotrimer core.
(A). Domain organizations of the α1 subunit of mammalian AMPK, and the Snf1 α subunit, the Sip2 β subunit, and the Snf4 γ subunit of S. cerevisiae SNF1. The protein kinase domain (KD) is in black, auto-inhibitory domain (AID) in blue, the regulatory sequence (RS) in red, and the carbohydrate binding module (CBM) in cyan. (B). Overall structure of the heterotrimer core of S. cerevisiae SNF1 [25], colored as in panel A. The KD-AID of Snf1 is missing in this structure. The structure figures were produced with PyMOL (www.pymol.org).
Crystal structures of the heterotrimer core (missing the KD-AID of the α subunit and an N-terminal segment of the β subunit) of S. pombe AMPK, S. cerevisiae SNF1, and mammalian AMPK have been reported recently (Fig. 1B) [24–27]. The structures show overall similarity for their equivalent regions, and demonstrate intimate interactions at the heterotrimer interface (Fig. 1B). The structure of S. cerevisiae SNF1 contains two additional regions, the CBM in the Sip2 β subunit and part of the RS in the Snf1 subunit (Fig. 1B) [25]. The RS (residues 392-518 of Snf1) was originally identified from yeast two-hybrid and biochemical assays, which indicated that this region of Snf1 interacted with both Snf4 and the Snf1 KD [11]. Mutation of Leu470 to Ser in the RS abolished interactions with Snf4 but had no effect on interactions with the KD in two-hybrid assays [11]. In the structure, residues 460-498 of the RS have tight interactions with Snf4 (Fig. 1B), and Leu470 has a major contribution to this interface (see below). Another study showed that deletion of residues 381-488 renders Snf1 independent of Snf4 for activation [28], although this segment also contains a part of the AID.
The fact that the RS can interact with both Snf4 and Snf1 KD lead to the model that it may help regulate SNF1 catalytic activity [11; 25]. Here we present biochemical data from GST pulldown experiments demonstrating strong, direct interactions between residues 450-500 of the RS and Snf4 in vitro, in agreement with the structural observations. On the other hand, the functional studies have so far only revealed small effects of these interactions on the activity of SNF1 in vivo. This suggests that residues 450–500 may be constitutively associated with Snf4, and the remaining segments of the RS, as well as the AID, may be involved in regulating SNF1 activity.
Materials and Methods
Protein expression and purification
Residues 450-500 of Snf1 were sub-cloned into the pGEX5X-1 vector (GE Healthcare) and over-expressed in E. coli BL21-Gold (DE3) cells (Stratagene) at 20 °C. The GST fusion protein, GST-RS, was purified by glutathione affinity and gel filtration chromatography. The heterotrimer cores of SNF1, with or without the CBM of Sip2, were co-expressed using a polycistronic expression system in the pET28a vector (Novagen) and purified by nickel affinity and gel filtration chromatography [25]. Both heterotrimers were concentrated to 10 mg/ml in a solution containing 50 mM Tris (pH 8.5), 150 mM NaCl, 5 mM DTT, and 5% (v/v) glycerol, and stored at −80 °C.
GST pull-down assays
45 µg of GST-RS fusion protein was incubated with 20 µl glutathione beads for 1 hour at 4 °C in a binding buffer containing 50 mM Tris (pH 8.5), 150 mM NaCl, 5 mM DTT, and 5% (v/v) glycerol. The heterotrimer core of SNF1 was added, and the mixture was incubated for 3 hours. The beads were centrifuged, resuspended with 1000 µl of the binding buffer to wash, centrifuged again, and the supernatant removed. Bound protein was eluted with the binding buffer supplemented with 20 mM reduced glutathione. Samples were resolved by SDS-PAGE and the proteins visualized using Coomassie blue or silver staining.
Expression of mutant Snf1 and Snf4 proteins in yeast
Centromeric plasmid pCE108 expressed Snf1 from its native promoter [29]. Mutations were introduced into pCE108 by site-directed mutagenesis [30] and confirmed by sequencing. Plasmids expressing wild-type and mutant Snf4 from its native promoter have been described previously [31]. Proteins were expressed in S. cerevisiae strain MCY5713 (W303-1A snf1Δ10 snf4Δ::kanMX4).
Immunoblot analysis
Yeast cultures were grown to exponential phase (A600 of 0.7) in selective synthetic complete (SC) medium containing 2% glucose. Cells were harvested by rapid filtration and either frozen in liquid nitrogen or resuspended in medum containing 0.05% glucose for 10 min, collected, and frozen. Cell extracts were prepared from two or three independent cultures as described [32]. SNF1 was partially purified on DEAE-Sepharose (GE Healthcare), separated by 8% SDS-PAGE and analyzed by immunoblotting using anti-phospho-Thr-172-AMPK (Cell Signaling Technologies) and anti-polyHistidine (Sigma) antibodies. ECL Plus (GE Healthcare) was used for visualization.
Results and Discussion
Analysis of the RS-Snf4 interface
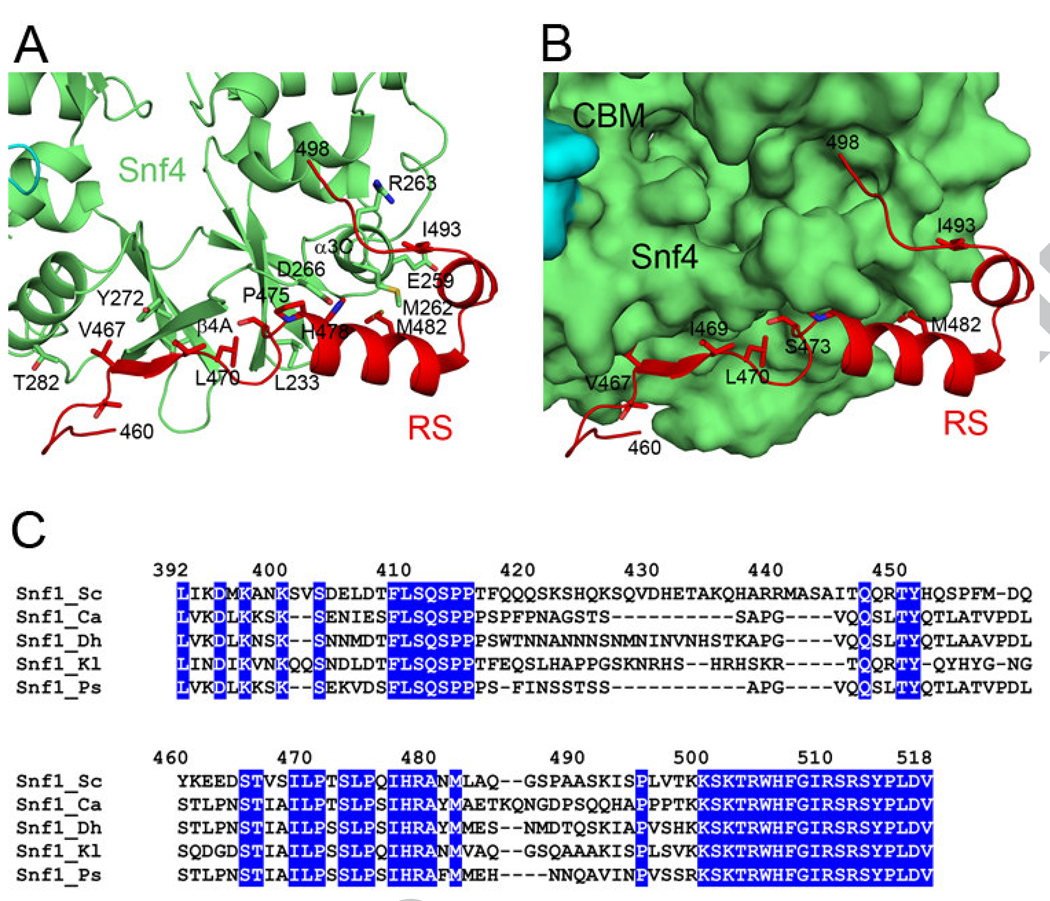
Residues 460-498 of the RS show extensive interactions with Snf4 in the structure of the S. cerevisiae SNF1 heterotrimer core (Fig. 1B) [25], which are predominantly hydrophobic in nature, but also include a small, anti-parallel β-sheet between residues 467-469 of RS and 270-275 (β4A) of Snf4 (Fig. 2A). This segment of RS lies in a shallow groove on the surface of the heterotrimer core (Fig. 2B), and approximately 1100 Å2 of its surface area is buried in the interface. A number of residues in this interface have >50 Å2 surface area burial, including Thr466, Val467, Ile469, Leu470, Ser473, Pro475, His478, Met482, and Ile493. Met482 of the RS interacts with a methionine residue in Snf4, Met262, which has 153 Å2 buried surface area in the interface (Fig. 2A). Other residues in Snf4 that contribute significantly to the buried surface are Ile233, Glu259, Arg263, Asp266, Val271, Tyr272, Thr273 and Thr282.
Figure 2. The RS-Snf4 interface in S. cerevisiae SNF1.
(A). Schematic drawing showing detailed interactions at the RS-Snf4 interface in the structure of the heterotrimer core of S. cerevisiae SNF1 [25]. (B). The Snf1 RS is located in a shallow groove on the surface of the heterotrimer core, colored as in Fig. 1B. (C). Sequence alignment of the Snf1 RS among fungal SNF1 enzymes, including S. cerevisiae (Sc), Candida albicans (Ca), Debaryomyces hansenii (Dh), Kluyveromyces lactis (Kl), and Pichia stiptis (Ps).
To assess the biochemical and functional importance of the RS-Snf4 interface, we created the V467R, L470S and M482A mutants in the RS, and the M262A mutant in Snf4. The L470S mutation was identified earlier to disrupt Snf1-Snf4 interactions in yeast two-hybrid assays [11]. As a control, we generated the E228K mutant of Snf4. This residue is located on the surface of Snf4, far away from the RS-Snf4 interface.
This segment of the RS (especially residues 465-482) is highly conserved among many fungal species (Fig. 2C), with the exception of S. pombe, which appears to support its potential functional importance. On the other hand, metazoan AMPKs appear to be missing most of this segment, suggesting that its function may be restricted to lower eukaryotes.
Biochemical evidence for direct interactions between Snf1 RS and heterotrimer core
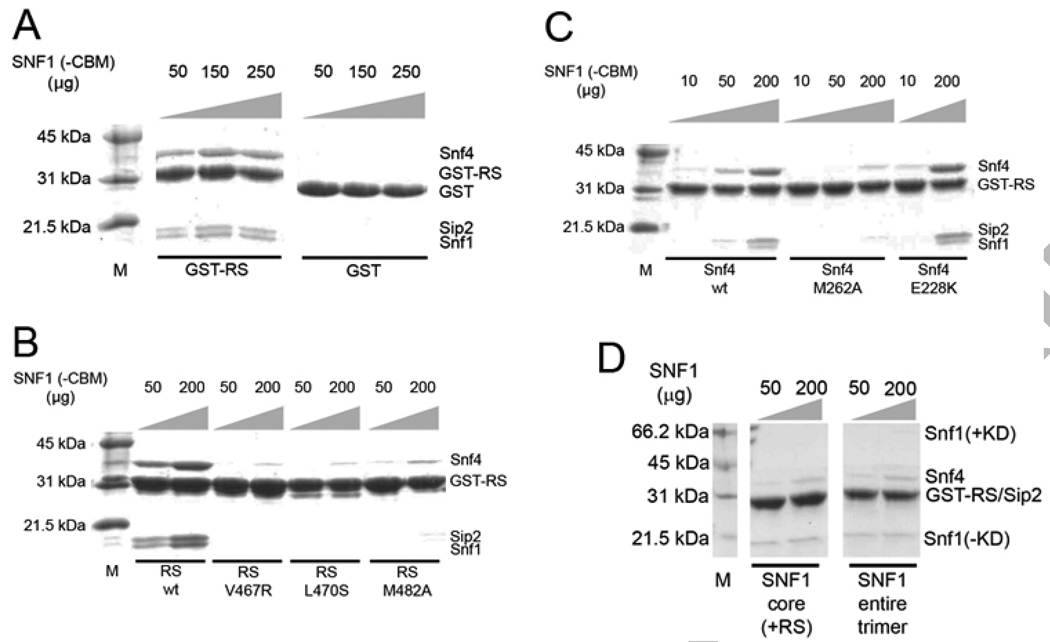
To obtain experimental evidence for direct interactions between the Snf1 RS and Snf4 in the heterotrimer core, we expressed residues 450-500 of Snf1 as a GST-fusion protein (GST-RS) and examined by GST pulldown assays the interactions between it and a heterotrimer core that is missing this segment of the RS and the CBM of Sip2. The GST-RS showed strong interactions with this heterotrimer core, as evidenced by visualization on a Coomassie-stained SDS gel (Fig. 3A). As a control, GST alone showed no interaction with the heterotrimer, even with a dose as high as 400 µg (Fig. 3A), demonstrating the specificity of the interactions.
Figure 3. Biochemical evidence for direct interactions between Snf1 RS and Snf4.
(A). GST pulldown experiments showing the interactions between wild-type GST-RS protein and the heterotrimer core of SNF1 that is missing the RS. GST alone showed no interactions. (B). Single-site mutations in the Snf1 RS, V467R, L470S, or M482A, are able to greatly reduce RS-Snf4 interactions. (C). A single-site mutation in Snf4, M262A, also disrupts RS binding. The control mutation, E228K, has no effect on the interactions. (D). GST pulldown experiments showing only weak interactions between wild-type GST-RS protein and the heterotrimer core (with RS) or the entire heterotrimer of SNF1.
We next examined the effects of single-site mutations in the RS-Snf4 interface on their interactions. GST-RS proteins carrying the V467R, L470S, or M482A mutation showed greatly reduced affinity for the heterotrimer core (Fig. 3B). Similarly, the heterotrimer core carrying the M262A mutation in Snf4 also showed significantly reduced interaction with wild-type GST-RS (Fig. 3C). In contrast, the control mutation in Snf4, E228K, had no effect on the interaction with GST-RS (Fig. 3C).
The pulldown experiments described above used a Sip2 variant that is missing its CBM. We repeated some of the pulldown assays utilizing a heterotrimer core that includes the CBM and obtained essentially the same results (data not shown). Therefore, the CBM is unlikely to be involved in regulating RS-Snf4 interactions. Overall, these results from the GST pulldown experiments are consistent with the structural data on the SNF1 heterotrimer core (Fig. 1B), and confirm that strong, direct interactions between the RS and Snf4 also occur in solution.
We also attempted to demonstrate the direct interactions between the CBM and the heterotrimer core (Fig. 1B) by GST pulldown experiments. The CBM of Sip2 was expressed and purified as a GST fusion protein, and its interactions with a heterotrimer core lacking the CBM were examined. We were not able to observe any binding between the two proteins by GST pulldown experiments (data not shown). Based on the structural information, the CBM-Snf4 interface is not as extensive as the RS-Snf4 interface, with about 650 Å2 of the surface area of CBM buried here, suggesting weaker interactions between the two proteins.
Evidence for only one strong binding site for Snf1 RS (residues 450-500) in the heterotrimer
As biochemical data suggest that the RS can interact with both the Snf1 KD and Snf4 [11], it may be expected that there are two binding sites for the RS in the SNF1 heterotrimer. Our structural and GST pulldown data have identified the binding site in Snf4. To examine whether there is a second binding site for the RS, possibly in the KD, we performed GST pulldown experiments using GST-RS protein (containing residues 450-500) and the entire SNF1 heterotrimer. A heterotrimer core, missing the KD-AID but containing the RS of Snf1, was used as the control.
As expected, weak interactions were observed in the pulldown experiments using the heterotrimer core (with RS, Fig. 3D). Interestingly, only weak interactions were observed in the pulldown experiments using the entire heterotrimer also (Fig. 3D). This demonstrates that there may be only one strong binding site for this segment of the RS, residues 450-500, in the SNF1 heterotrimer, and this binding site is located in Snf4. It could be possible that binding to Snf1 KD is mediated by the C-terminal segment of RS, residues 496-518, as suggested by earlier data [11]. We were however unable to express the entire RS as a GST fusion protein to carry out pulldown experiments.
Functional effects of mutations in the RS-Snf4 interface
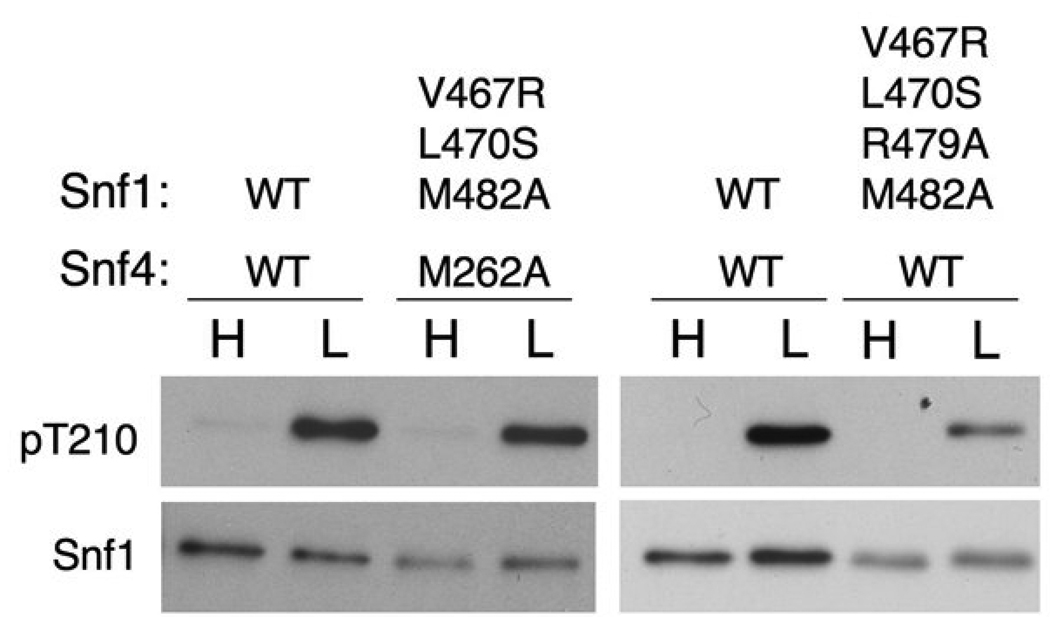
To test the functional significance of the interactions between the RS and Snf4 in vivo, we introduced mutations in this interface into SNF1 and SNF4. The mutant proteins were expressed from their native promoters on centromeric plasmids in snf1Δ snf4Δ S. cerevisiae cells. Cultures were grown to exponential phase in medium containing high (2%) glucose, an aliquot was collected, and cells were then subjected to glucose depletion by shifting to medium containing 0.05% glucose for 10 min. Cells were harvested by rapid filtration to maintain the phosphorylation state of Snf1, and extracts were prepared and assayed for SNF1 catalytic activity and phosphorylation of Thr210. Cells expressing the V467R, L470S, or M482A mutant of Snf1 and either wild-type Snf4 or the M262A mutant showed no defect in glucose inhibition of SNF1 or activation of SNF1 in response to glucose depletion (data not shown). We then introduced all four mutations into the same cell, involving a triple mutant (V467R/L470S/M482A) of Snf1, and still observed no defect in regulation of phosphorylation or SNF1 activity under the condition tested (Fig. 4 and data not shown). Introduction of a further mutation into Snf1, R479A, also failed to produce a significant phenotype (Fig. 4).
Figure 4. Effect of RS mutations on Snf1 phosphorylation in vivo.
Wild-type (WT) and mutant Snf1 and Snf4 proteins, as indicated, were expressed in snf1Δ snf4Δ cells. Cells were grown to mid-log phase in selective SC plus 2% (high, H) glucose, and an aliquot was harvested by rapid filtration. Another aliquot was harvested, resuspended in SC plus 0.05% (low, L) glucose for 10 min, and harvested by rapid filtration. Partially purified SNF1 was analyzed by immunoblotting with anti-phospho-T172-AMPK antibody to detect phosphorylated Thr-210 (pT210) of Snf1 and, on a separate blot, with anti-polyhistidine antibody to detect Snf1 protein.
In yeast, SNF1 activity is inhibited in high glucose conditions, and is activated when the cells are shifted to low glucose. Snf4 is required for this activation in response to glucose depletion [29; 33; 34]. However, deletion of C-terminal residues of Snf1, from 309 or 392 to the C-terminus or residues 381-488, renders it independent of Snf4 for activation. This suggests that Snf4 may counteract Snf1 auto-inhibition by its C-terminal segment [11; 28; 29], and it also implies that there may be interactions between the two subunits, which were shown by yeast two-hybrid assays [11]. While our structural and biochemical data demonstrated direct and strong interactions between residues 450-500 of RS and Snf4 (Fig. 3A) [11; 25], the functional studies appear to suggest that disruption of this RS-Snf4 interface has only a small effect on SNF1 activity.
Overall, the experimental data suggest that the segment of RS that we studied here, residues 450-500, may have constitutive interactions with Snf4, and may not be involved in regulating the activity of the SNF1 heterotrimer. At the same time, the functional importance of the entire RS (residues 392-518) was demonstrated by earlier studies [11]. Further experiments are needed to establish the molecular mechanism for how this entire RS contributes to the regulation of SNF1 activity.
Acknowledgements
We thank David Stapleton for helpful discussions. This research is supported in part by NIH grants DK67238 (to LT) and GM34095 (to MC). GAA was also supported by an NIH training program in molecular biophysics (GM08281).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Ann. Rev. Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 2.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metabolism. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Kemp BE, Stapleton D, Campbell DJ, Chen Z-P, Murthy S, Walter M, Gupta A, Adams JJ, Katsis F, van Denderen B, Jennings IG, Iseli T, Michell BJ, Witters LA. AMP-activated protein kinase, super metabolic regulator. Biochem. Soc. Trans. 2003;31:162–168. doi: 10.1042/bst0310162. [DOI] [PubMed] [Google Scholar]
- 4.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell. Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 5.Sanz P. Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem. Soc. Trans. 2003;31:178–181. doi: 10.1042/bst0310178. [DOI] [PubMed] [Google Scholar]
- 6.Rudolph MJ, Amodeo GA, Bai Y, Tong L. Crystal structure of the protein kinase domain of yeast AMP-activated protein kinase Snf1. Biochem. Biophys. Res. Commun. 2005;337:1224–1228. doi: 10.1016/j.bbrc.2005.09.181. [DOI] [PubMed] [Google Scholar]
- 7.Nayak V, Zhao K, Wyce A, Schwartz MF, Lo WS, Berger SL, Marmorstein R. Structure and dimerization of the kinase domain from yeast Snf1, a member of the Snf1/AMPK protein family. Structure. 2006;14:477–485. doi: 10.1016/j.str.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Crute BE, Seefeld K, Gamble J, Kemp BE, Witters LA. Functional domains of the a1 catalytic subunit of the AMP-activated protein kinase. J. Biol. Chem. 1998;273:35347–35354. doi: 10.1074/jbc.273.52.35347. [DOI] [PubMed] [Google Scholar]
- 9.Pang T, Xiong B, Li J-Y, Qiu B-Y, Jin G-Z, Shen J-K, Li J. Conserved a-helix acts as autoinhibitory sequence in AMP-activated protein kinase a subunits. J. Biol. Chem. 2007;282:495–506. doi: 10.1074/jbc.M605790200. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Jiao Z-H, Zheng L-S, Zhang Y-Y, Xie S-T, Wang Z-X, Wu J-W. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459:1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 11.Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 12.Jiang R, Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iseli TJ, Walter M, van Denderen BJW, Katsis F, Witters LA, Kemp BE, Michell BJ, Stapleton D. AMP-activated protein kinase b subunit tethers a and g subunits via its C-terminal sequence (186–270) J. Biol. Chem. 2005;280:13395–13400. doi: 10.1074/jbc.M412993200. [DOI] [PubMed] [Google Scholar]
- 14.Polekhina G, Gupta A, Michell BJ, van Denderen B, Murthy S, Feil SC, Jennings IG, Campbell DJ, Witters LA, Parker MW, Kemp BE, Stapleton D. AMPK beta subunit targets metabolic stress sensing to glycogen. Curr. Biol. 2003;13:867–871. doi: 10.1016/s0960-9822(03)00292-6. [DOI] [PubMed] [Google Scholar]
- 15.Hudson ER, Pan DA, James J, Lucocq JM, Hawley SA, Green KA, Baba O, Terashima T, Hardie DG. A novel domain in AMP-activated protein kinase causes glycogen storage bodies similar to those seen in hereditary cardiac arrhythmias. Curr. Biol. 2003;13:861–866. doi: 10.1016/s0960-9822(03)00249-5. [DOI] [PubMed] [Google Scholar]
- 16.Polekhina G, Gupta A, van Denderen BJ, Feil SC, Kemp BE, Stapleton D, Parker MW. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13:1453–1462. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Koay A, Rimmer KA, Mertens HD, Gooley PR, Stapleton D. Oligosaccharide recognition and binding to the carbohydrate binding module of AMP-activated protein kinase. FEBS Lett. 2007;581:5055–5059. doi: 10.1016/j.febslet.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 18.Scott JW, van Denderen BJ, Jorgensen SB, Honeyman JE, Steinberg GR, Oakhill JS, Iseli TJ, Koay A, Gooley PR, Stapleton D, Kemp BE. Thienopyridone drugs are selective activators of AMP-activated protein kinase beta1-containing complexes. Chem. Biol. 2008;15:1220–1230. doi: 10.1016/j.chembiol.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 20.Kemp BE. Bateman domains and adenosine derivatives form a binding contract. J. Clin. Investig. 2004;113:182–184. doi: 10.1172/JCI20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Investig. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudolph MJ, Amodeo GA, Iram S, Hong S-P, Pirino G, Carlson M, Tong L. Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding. Structure. 2007;15:65–74. doi: 10.1016/j.str.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Day P, Sharff A, Parra L, Cleasby A, Williams M, Horer S, Nar H, Redemann N, Tickle I, Yon J. Structure of a CBS-domain pair from the regulatory g1 subunit of human AMPK in complex with AMP and ZMP. Acta Cryst. 2007;D63:587–596. doi: 10.1107/S0907444907009110. [DOI] [PubMed] [Google Scholar]
- 24.Townley R, Shapiro L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science. 2007;315:1726–1729. doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- 25.Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Sacharyomyces cerevisiae AMPK homolog SNF1. Nature. 2007;449:492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- 26.Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, Martin SR, Carling D, Gamblin SJ. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 27.Jin X, Townley R, Shapiro L. Structural insight into AMPK regulation: ADP comes into play. Structure. 2007;15:1285–1295. doi: 10.1016/j.str.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Leech A, Nath N, McCartney RR, Schmidt MC. Isolation of mutations in the catalytic domain of the snf1 kinase that render its activity independent of the snf4 subunit. Eukaryot. Cell. 2003;2:265–273. doi: 10.1128/EC.2.2.265-273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Celenza JL, Carlson M. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell. Biol. 1989;9:5034–5044. doi: 10.1128/mcb.9.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–571. doi: 10.2144/97234bm01. 574. [DOI] [PubMed] [Google Scholar]
- 31.Momcilovic M, Iram SH, Liu Y, Carlson M. Roles of the glycogen-binding domain and Snf4 in glucose inhibition of SNF1 protein kinase. J. Biol. Chem. 2008;283:19521–19529. doi: 10.1074/jbc.M803624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedbacker K, Hong S-P, Carlson M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell Biol. 2004;24:8255–8263. doi: 10.1128/MCB.24.18.8255-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celenza JL, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 34.Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]