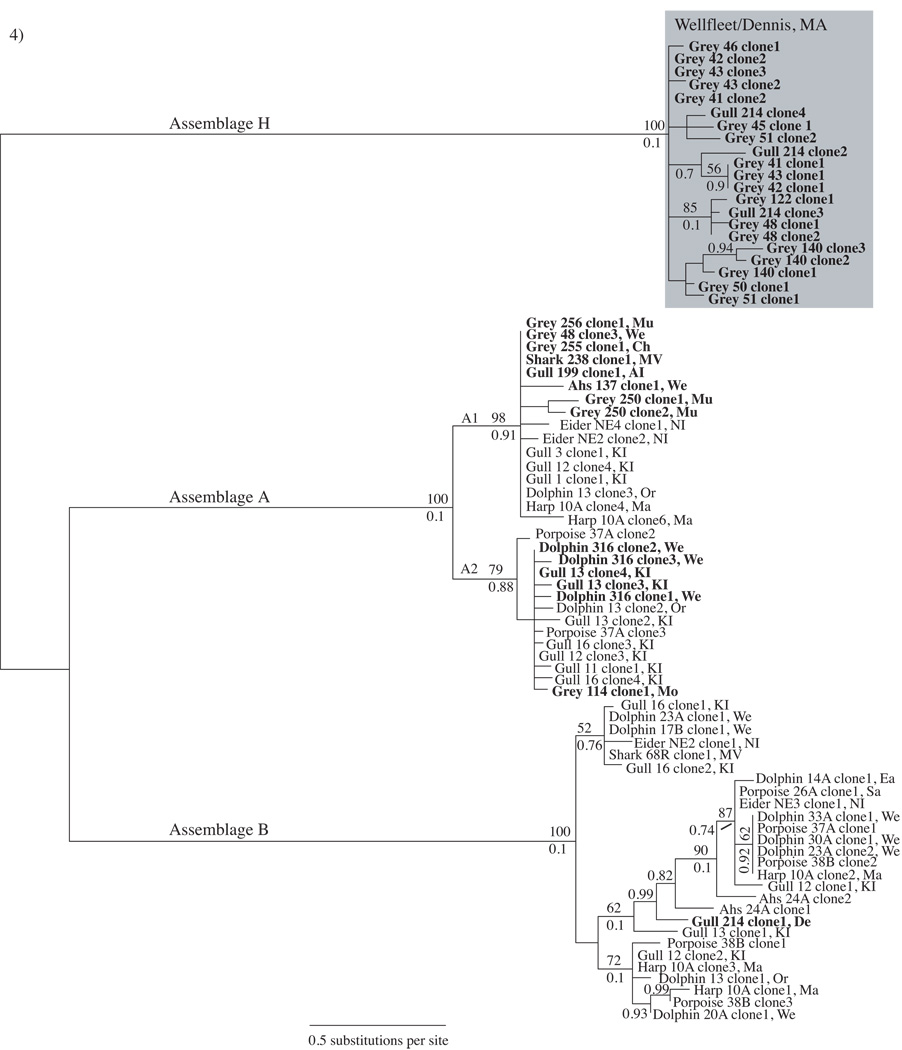
Fig. 4.
Giardia duodenalis glutamate dehydrogenase (gdh) Bayesian tree derived from east coast marine animal fecal samples. The gene tree is a consensus of 75,000 sampled trees. Boldface indicates sequences generated from this study. Grey, Ahs, Phs and harp refer to grey seal, Atlantic harbor seal, Pacific harbor seal and harp seal, respectively. Boostrap values > 50 are shown above branches and posterior probabilities > 0.70 are shown below branches unless a boostrap value is not present. The shaded box highlights Assemblage H sequences derived from 10 grey seals from Jeremy Point, Wellfleet, Massachusetts (MA, USA) and one gull from Dennis, MA. Sampling site abbreviations are included with each sample: KI, Kent Island, Canada; We, Wellfleet, MA; De, Dennis, MA; Sa, Sandwich, MA; Or, Orleans, MA; Ch, Chatham, MA; Mo, Monomoy, MA; Mu, Muskeget, MA; MV, Martha’s Vineyard Island, MA; NI, Nantucket Island, MA; AI, Appledore Island, Maine; Ma, Marion, MA. A1 and A2 denote subgroups of Assemblage A.