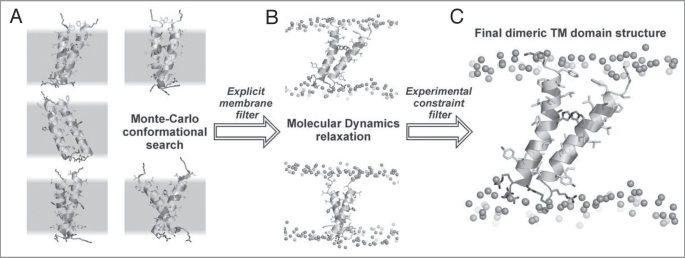
Figure 1.
Spatial structure elucidation of dimeric TM domain of bitopic protein (exemplified by proapoptotic protein BNip3) using computer simulations techniques. (A) Collection of rough models via Monte Carlo conformational search in implicit membranes. Set of possible structure candidates having minimal energy is presented. Peptides are given in ribbon and stick representations. Positions of implicit membrane are selected by gray hatching. (B) Results of MD-relaxation of the MC structures in full-atom hydrated bilayer. Only structures stable in the membrane during MD are selected. Membranes are delineated by position of phosphorus atoms (shown as spheres). Other details were skipped for clarity. (C) The resulting structure selected via comparison with experimental information about dimer interface.