Figures 2.
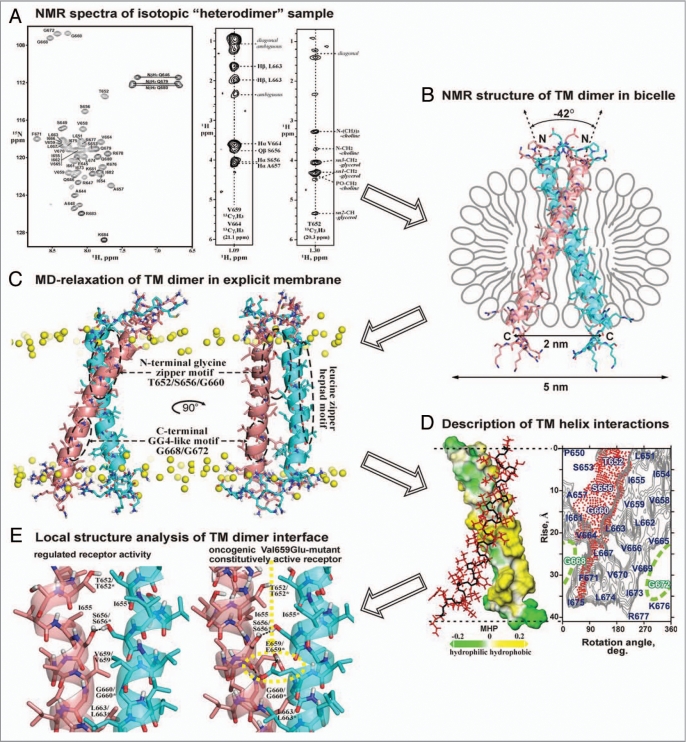
Spatial structure elucidation of dimeric TM domain of the bitopic protein (exemplified by receptor tyrosine kinase ErbB2)130 using with the aid of heteronuclear solution NMR in lipid bicelles and MD-relaxation in explicit lipid bilayer. (A) Acquisition of NMR spectra of isotopic “heterodimer,” consisting of 13C/15N-isotope labeled and natural abundance ErbB2 TM fragments (residues 641–685) embedded into DMPC/DHPC lipid bicelles. From left to right, 1H-15N HSQC spectrum with amide backbone resonance assignments, two representative 2D strips from the 3D 13C F1-filtered/F3-edited-NOESY spectrum with intermolecular protein-protein and protein-lipid NOE contacts are presented. (B) Determination of high-resolution spatial structure of the right-handed ErbB2 TM homodimer in lipid bicelle using NMR-derived restraints. The obtained N-terminal association mode of the ErbB2 TM dimer via N-terminal dimerization motif corresponds to the receptor active state. (C) MD-relaxation of the ErbB2 TM homodimer in hydrated explicit DMPC lipid bilayer with imposed NMR-derived constraints. Yellow balls show phosphorus atoms of lipid heads. The spatial locations of the three characteristic dimerization motifs of ErbB2tm are marked by dashed ovals. (D) Analysis of interacting surfaces of the ErbB2 TM helices. In left, hydrophobic and hydrophilic (polar) surfaces of one TM helix in the homodimer colored in yellow and green according to the molecular hydrophobicity potential (MHP).132 The second monomer of the dimer is shown with red side chains. In right, hydrophobicity map for ErbB2 TM helix surface with contour isolines encircling hydrophobic regions with high values of MHP is presented with red-point area indicating the helix packing interface via N-terminal glycine zipper motif T652xxxS656xxxG660. The residues composing C-terminal unemployed dimerization GG4-like motif G668xxxG672 are highlighted in green. (E) Local structure analysis of intra and intermolecular interactions in the ErbB2 TM dimer. Comparison of intermonomeric hydrogen bonding (black dotted lines) in the TM helix-helix interface of ErbB2 and its constitutively active Val659Glu-mutant is presented.