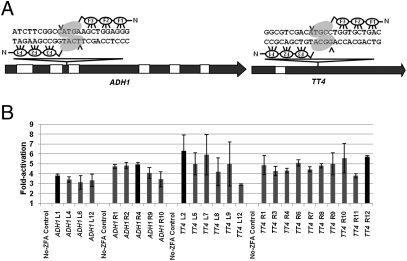
Fig. 1.
OPEN ZFN target sites in the ADH1 and TT4 coding sequences. (A) The gene models of Arabidopsis ADH1 and TT4 loci are shown as block arrows. Black rectangles represent exons; white rectangles represent introns. The positions of OPEN ZFN target sites are indicated by triangles. Each engineered ZFA consists of three zinc fingers (F1, F2, F3), which together recognize a 9-bp target site. The two target sites are separated by 6-bp spacer sequences. Binding of the ZFAs to the target sequences enables the FokI nuclease monomers (indicated as gray ovals) to dimerize and cleave within the spacer. The sites of cleavage in the target sequences are indicated by black arrowheads. (B) ZFAs generated by OPEN were tested for their activity in bacterial two-hybrid (B2H) assays. The ZFAs were tested for their ability to bind target sequences in bacteria and activate transcription of a downstream lacZ reporter gene. Fold activation of each ZFA relative to its negative control, which does not express the ZFA, is plotted on the y axis. Multiple ZFAs generated by OPEN were tested for each of the ADH1 and TT4 left and right target sites. ZFAs with activities indicated by black bars were used in subsequent tests. Error bars denote SD; n = 3.